Method for detecting hepatitis B virus DNA and G1896A mutation thereof and kit
A hepatitis B virus, G1896A technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of prolonging the running time of PCR programs, etc., achieve beautiful and stable amplification curves, reliable detection results, and benefit Promoted app performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Embodiment 1: Extraction of hepatitis B virus DNA in clinical blood samples
[0098] Take 50 μl of serum from clinical blood samples into a high-pressure 1.5ml centrifuge tube, add 25 μl of extraction solution A (40% PEG-8000), shake and mix for 5 seconds, centrifuge at 12,500 g for 5 minutes; discard the supernatant, add 25 μl of extraction Solution B (0.25M NaOH), shake for 15s to dissolve the precipitate as much as possible, lyse at 96°C for 10min; add 25μl of extract solution C (0.4M Tris-HCl buffer, pH6.4), mix well, 96°C for 10min; centrifuge at 12,500g After 15 minutes, 5 μl of the supernatant was taken for PCR amplification.
Embodiment 2
[0099] Embodiment 2: PCR amplification of target nucleic acid
[0100] Add a single serving of PCR reaction mixture into a PCR reaction tube, then add 0.2 μl of enzyme mixture, and finally add 5 μl of template (template to be tested or negative and positive control), mix thoroughly and centrifuge slightly (about 5 Second). Then put the reaction tube into a fluorescent PCR instrument (a PCR instrument that can detect FAM and HEX), and amplify according to the following conditions: 50°C for 120 seconds (1 cycle) → 95°C for 180 seconds (1 cycle) → 95°C for 10 seconds seconds, 56°C for 30 seconds (40 cycles), where 56°C is the temperature for detecting fluorescence.
[0101] The PCR amplification system used includes: 2.5 μl 10x PCR buffer (850 mM KCl, 400 mM Tris-Cl, pH8.9, 50% v / v glycerol), 0.2 μl enzyme mixture, 0.2 μl dUTPs (containing dATP, dUTP, dGTP , dCTP each 25mM), forward and reverse two primers (SEQ ID NO: 1 and SEQ ID NO: 2, 100 μ M) each 0.1 μ l, wild-type and mut...
Embodiment 3
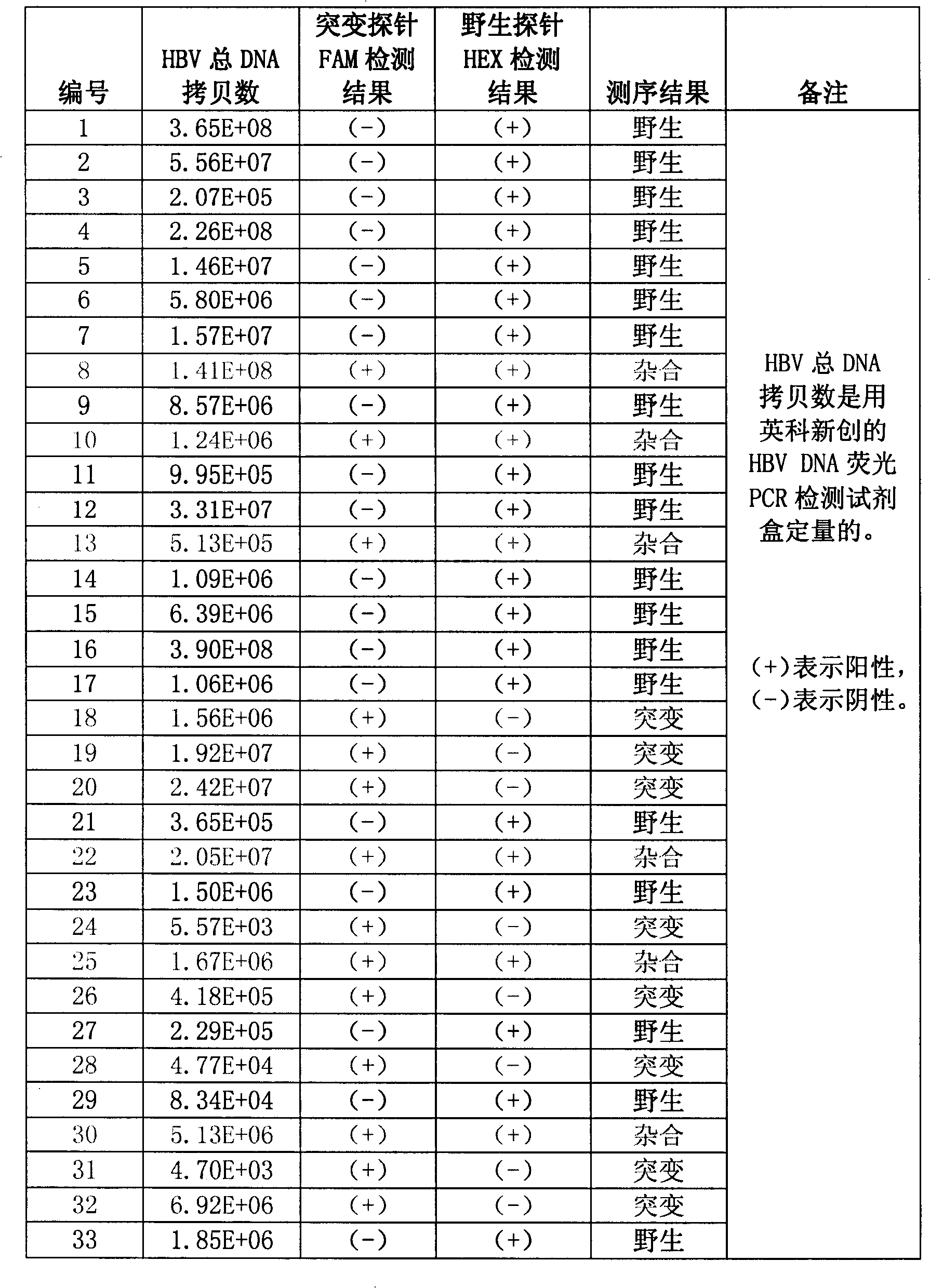
[0102] Example 3: Hepatitis B virus DNA G 1896 Sensitive and specific detection of A mutations
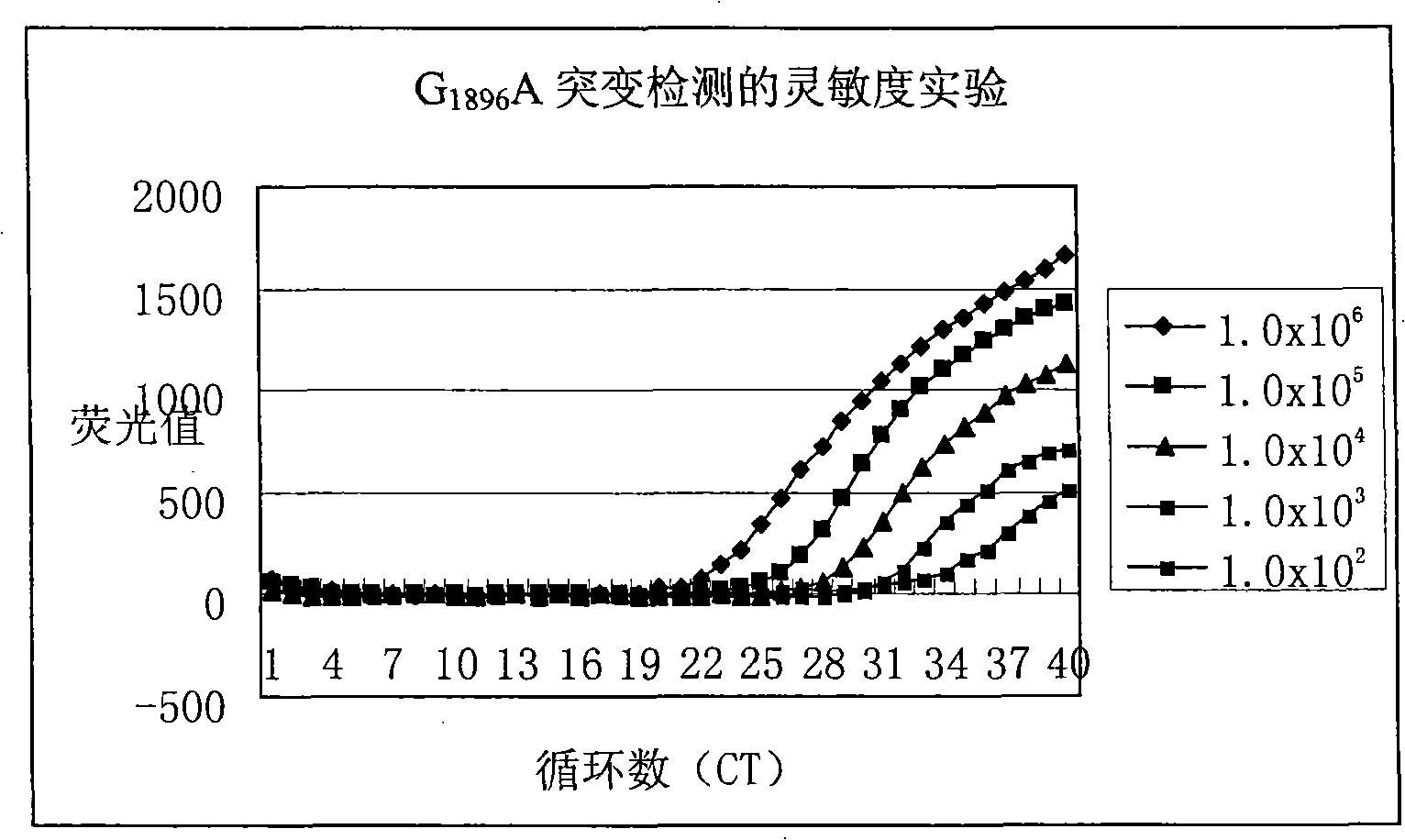
[0103] with G 1896 A plasmid (1.0x10) of a mutated hepatitis B virus DNA fragment 6 copies / ml-1.0x10 2 copies / ml) is template, use reagent of the present invention to hepatitis B disease
[0104] Toxic DNAG 1896 The sensitivity of A mutation is detected, and the detection results can be found in figure 1 . from figure 1 As can be seen from the results, the method and kit of the present invention can at least detect that the template concentration is only 1.0x10 2 copies / ml of G 1896 A mutated hepatitis B virus DNA. In other words, the sensitivity of the present invention can fully meet the clinical needs.
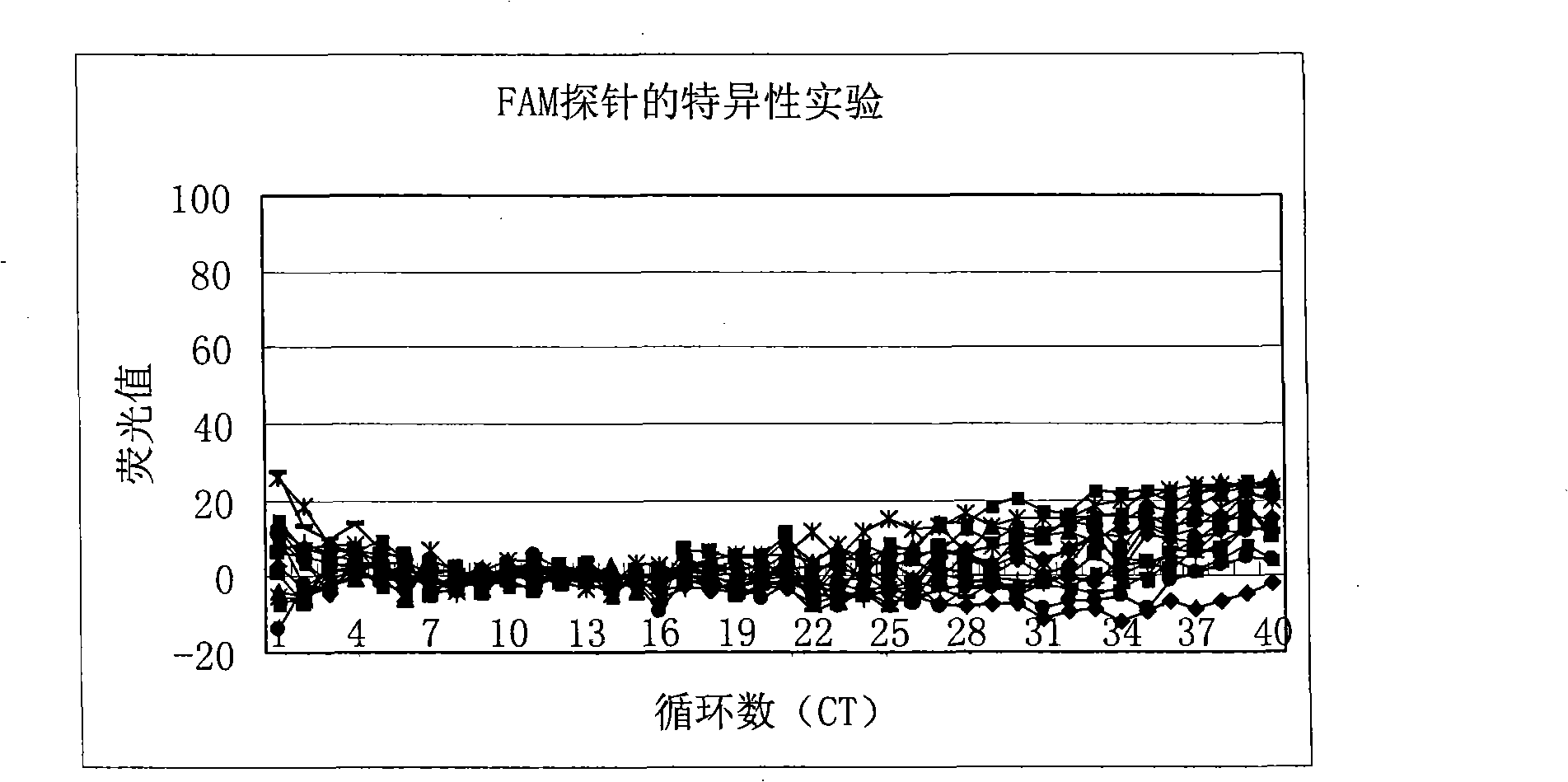
[0105] Then wild-type hepatitis B virus DNA (about 1.0×10 6 copies / ml) is a template, and the specificity of the FAM-labeled mutant probe is detected using the reagent of the present invention, and the detection results can be found in figure 2 . from figure 2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com