Biological sequence analysis method based on gap spectrum
A biological sequence and analysis method technology, applied in the field of non-comparison analysis, can solve the problems of not being able to use structure and function, and not being able to meet high speed and high sensitivity at the same time, and achieve the effect of performance improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 DNA sequence analysis based on gap spectrum
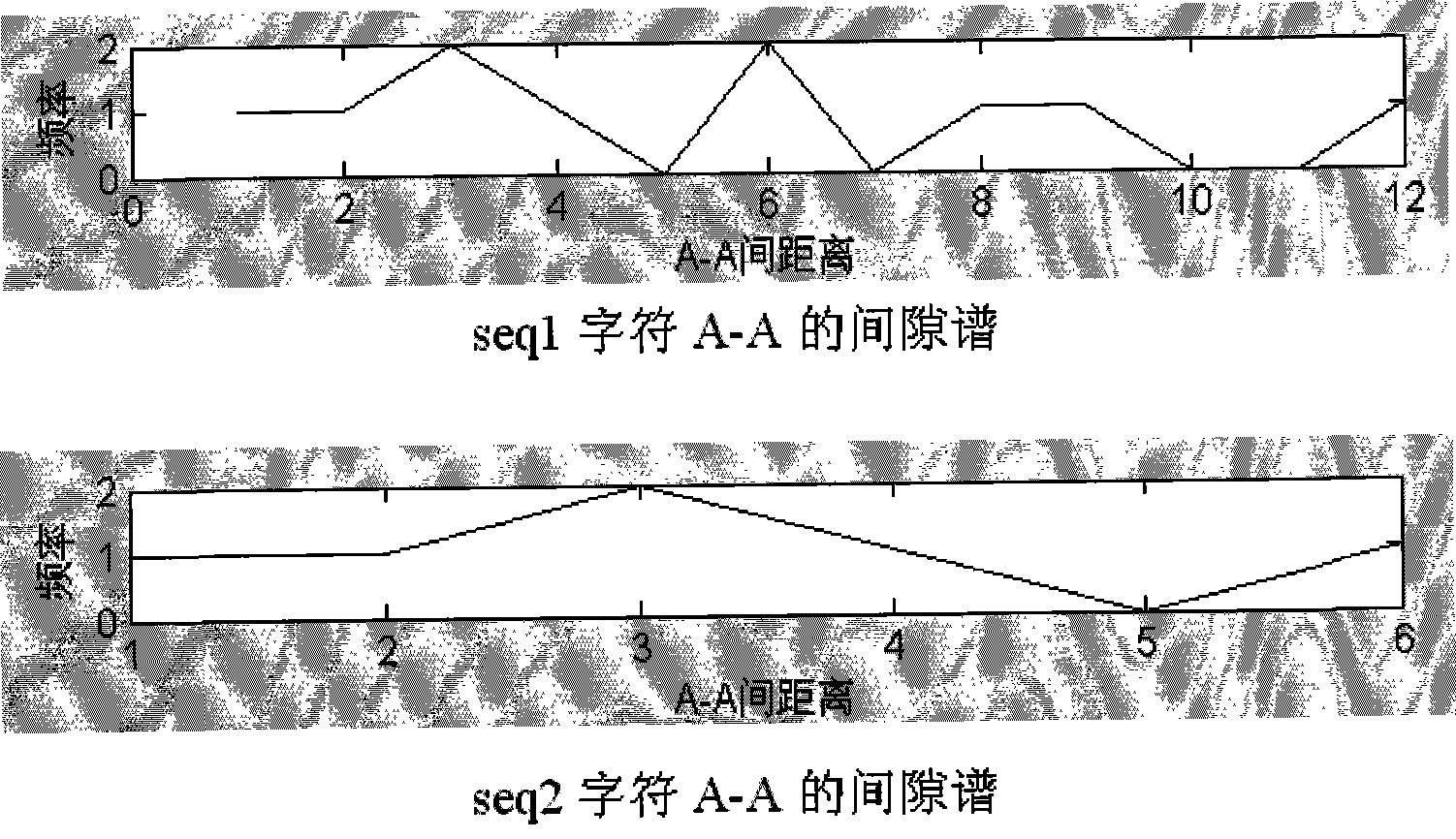
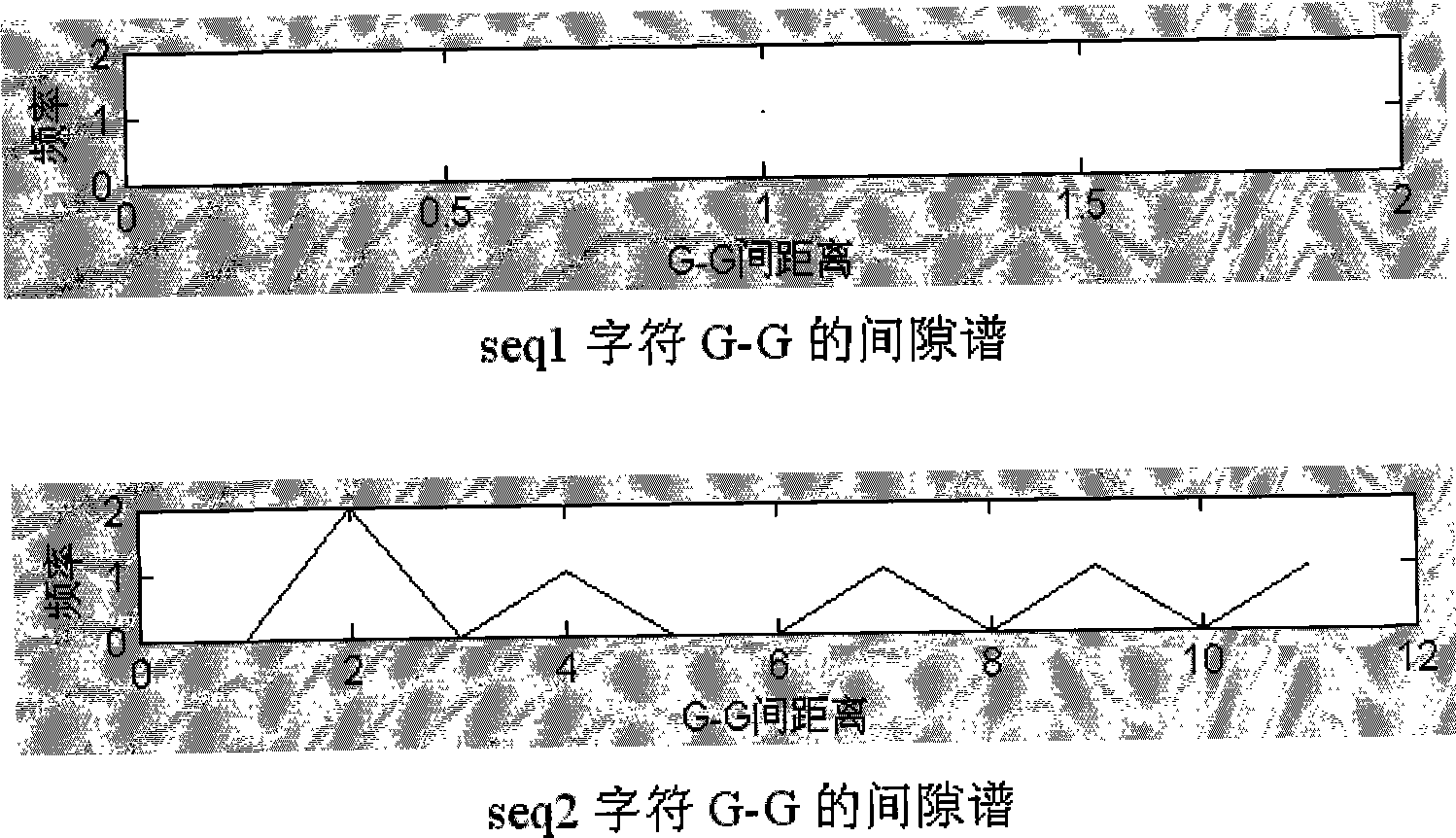
[0036] The subsequence (seq1) of the DNA sequence (SEQ1) with a full length of 20 is marked with one-dimensional coordinates.
[0037] A A A A G G C C T A C A A T T A C T C T DNA sequence with a full length of 20 (negative strand direction)
[0038] The DNA sequence (SEQ1) with a full length of 20 is marked with one-dimensional coordinates along its minus-strand direction. The coordinate identification starts at the 4th base (identified as 1), ends at the penultimate base 5 (identified as 13), and finally obtains a DNA subsequence (seq1) with a length of 13 and a one-dimensional coordinate identification. The identification results are as follows:
[0039] A G G C C T A C A A T T A DNA subsequence of length 13
[0040] 1 2 3 4 5 6 7 8 9 10 11 12 13 A DNA subsequence of length 13 identified by one-dimensional coordinates
[0041] Taking character A as an example, find the coordinates of the second, third, and last...
Embodiment 2
[0151] This embodiment introduces a new feature to the two DNA sequences on the basis of Embodiment 1, that is, for the frequency data of the gap spectrum (Table 16) in Embodiment 1, calculate the gap between the characters when the corresponding frequency is maximum in the gap spectrum. The calculation results are shown in Table 21.
[0152] Table 21 The distance between characters at the maximum frequency
[0153] frequency statistics A-A A-C A-G A-T C-A C-C C-G C-T G-A G-C G-G G-T T-A T-C T-G T-T Frequency in seq1
characters at max
spacing
3,6
3,4,7
2
2,5
5
3,4
/
7
7
2
/
9
2,3,4
,7
2
/
5,6 Frequency in seq2
characters at max
spacing
3
5,9
4,6,8...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com