Zinc finger nuclease-mediated homologous recombination
A technology of homologous recombination and nuclease, applied in the direction of recombinant DNA technology, hydrolase, peptide, etc., can solve the problems that cannot be widely used
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0353]Example 1 - Design and Generation of Targeting Vectors
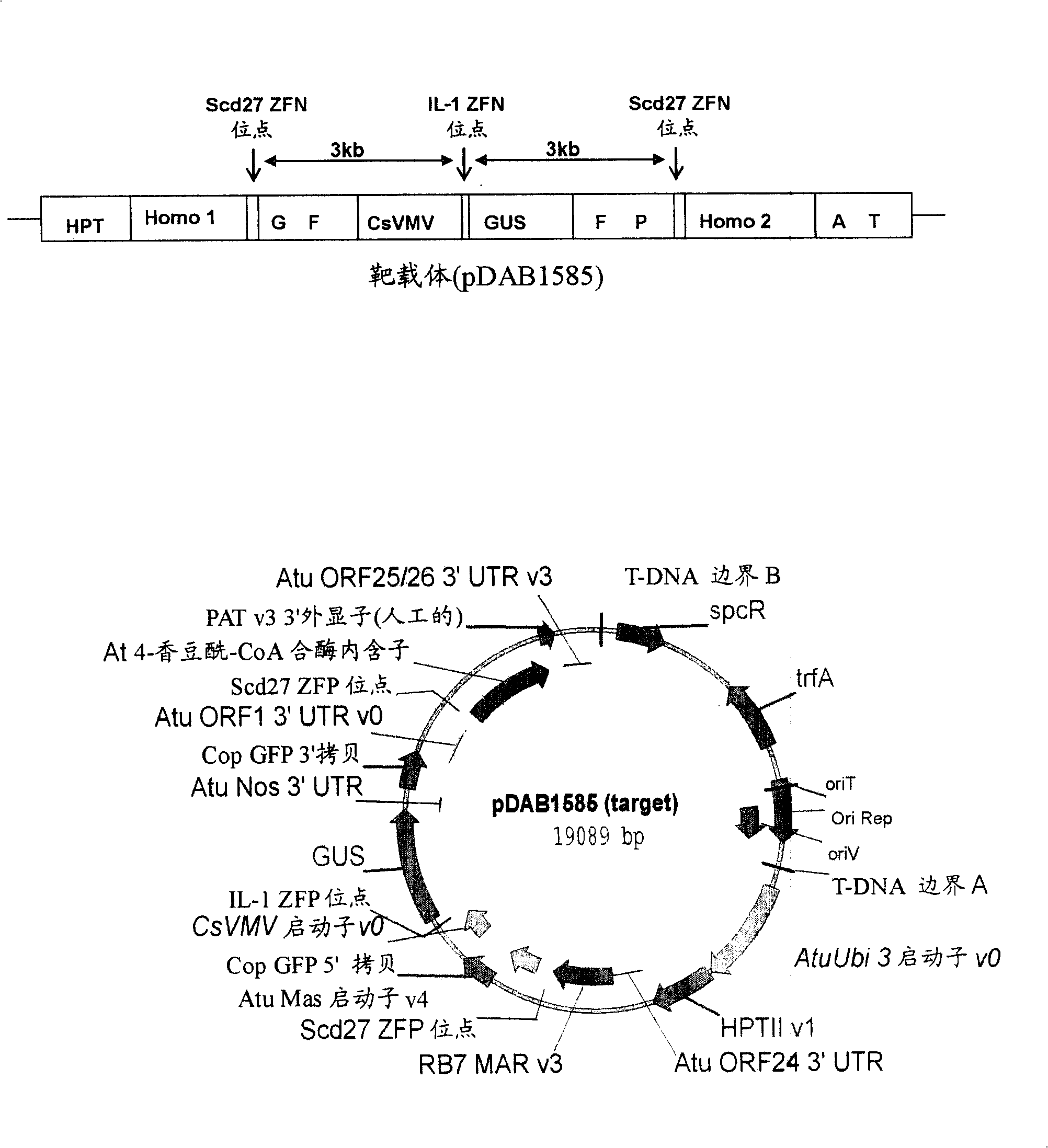
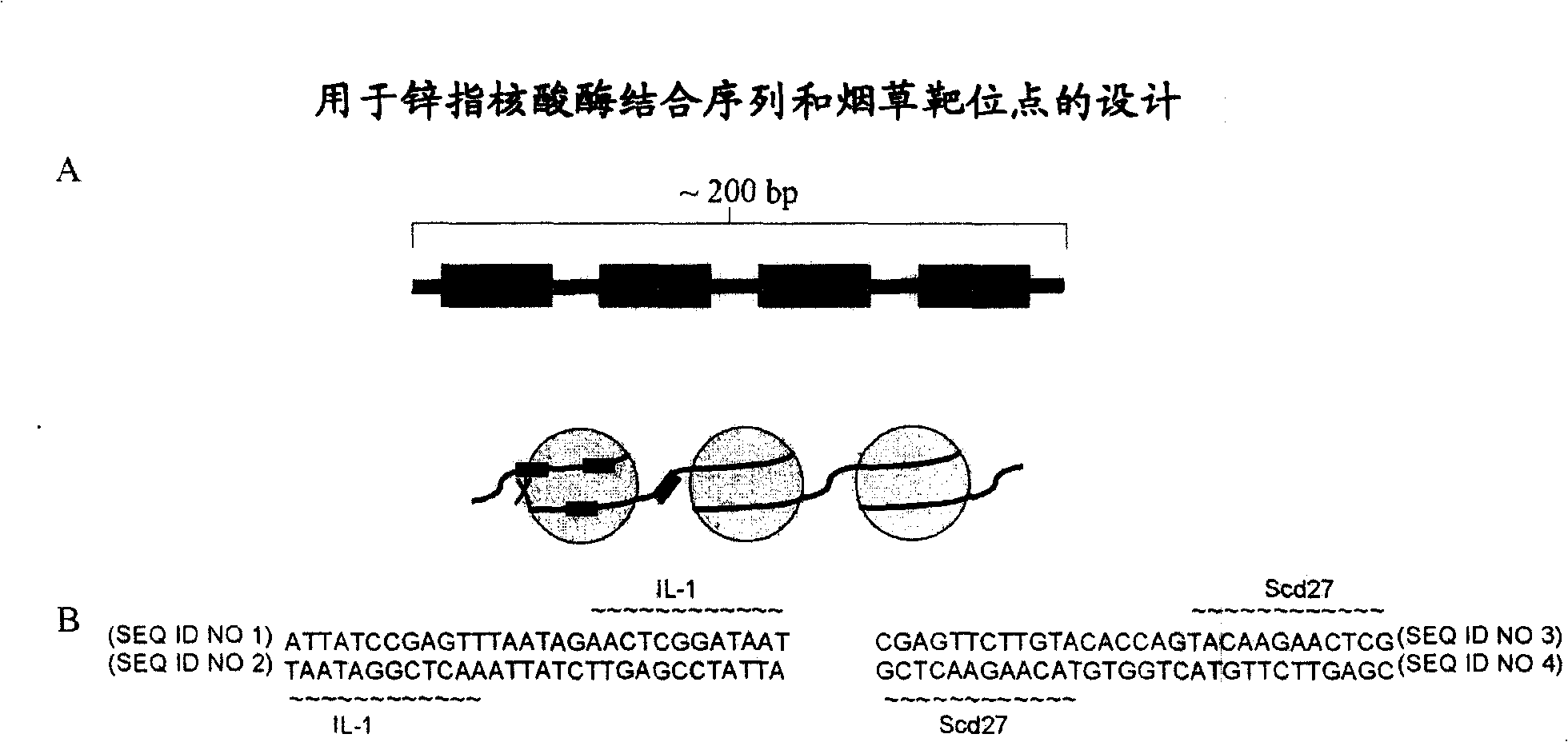
[0354] A. Overall Structure of the Target Sequence
[0355] Target constructs for tobacco (dicots) include the following 7 building blocks, such as figure 1 Shown: i) hygromycin phosphotransferase (HPT) expression cassette, which comprises the drive by Agrobacterium tumefaciens (A.tumifaciens) open reading frame-24 (orf-24) 3' untranslated region (UTR) (Gelvin etc., 1987, EP222493) the Escherichia coli HPT gene (Waldron et al., 1985, Plant Mol.Biol.18:189-200) terminated Arabidopsis (A.thaliana) ubiquitin-3 (ubi-3) promoter (Callis et al., 1990, J.Bio.Chem.265:12486-12493); ii) homologous sequence-1, which comprises the matrix attachment region (MAR) of tobacco (N. tabacum) RB7 (Thompson et al., 1997, WO9727207) ; iii) 5' green fluorescent protein (GFP) gene fragment (Evrogen Joint Stock Company, Moscow, Russia), which is expressed by the modified Agrobacterium tumefaciens mannopine synthase (Δmas) promoter (Pe...
Embodiment 2
[0374] Example 2 - Generation of transgenic cell lines with integrated target sequences
[0375] Two different suspension cultures of tobacco cells in which the target sequence of Example 1 was stably integrated via Agrobacterium transformation were used. The first culture, called NT1, was obtained from Arnold Bendich (University of Washington, Seattle, WA, USA). The culture proliferated to 15-20 μ diameter cells in 20-30 cell clusters with a doubling time of approximately 48 hours. NT1 cell suspension cultures were maintained in media containing the following components: MS basal salts (PhytoTechnology Labs M524), 137.4 mg / L K 2 HPO 4 , 30g / L sucrose, 2.22mg / L2,4-D, 1mg / L thiamine hydrochloride, 100mg / L myo-inositol and 0.5g / L MES, pH 5.7. NT1 cells were subcultured every 7 days by adding 40 mL of fresh MS-based medium to 1 mL of packed cell volume (PCV).
[0376] The second tobacco cell culture used (which was called BY2) was obtained from Jun Ueki (Japan Tobacco, Iwata,...
Embodiment 3
[0381] Example 3 - Screening and Characterization of Targeted Transgenic Events
[0382] Hygromycin resistance transgenic events (as described in Example 2) generated from transformation of targeting vectors into BY2 or NT1 tobacco cell cultures were analyzed as follows.
[0383] Initial analyzes to screen for these transgenic events included GUS expression analysis to demonstrate target sequence accessibility, PCR analysis of partial and full-length target sequences to confirm the presence and integrity of the target vector, and Southern blot analysis to determine integrated copy number of the target sequence. A subset of transgenic events showing GUS expression contained a single copy of the full-length target sequence; they were selected for reconstitution of suspension cultures to generate target lines for subsequent retransformation. These reconstructed target lines also underwent further characterization, which included more comprehensive Southern blot analysis, sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com