Set of yeast cells, method of identifying target candidate molecule, method of analyzing action mechanism and screening method
A technology of yeast cells and target candidates, which is applied in the field of yeast cell sets, and can solve the problems of decreased drug sensitivity of yeast cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0078] The specific operation method in the above-mentioned preparation method can be followed, for example, in Molecular Cloning (molecular cloning): A Laboratory Manual, Cold Spring Harbor, NY, Methods in Yeast Genetics (yeast genetics method): A Cold Spring Harbor Laboratory Course Manual, 2000 Edition, etc. The method described in the operation.
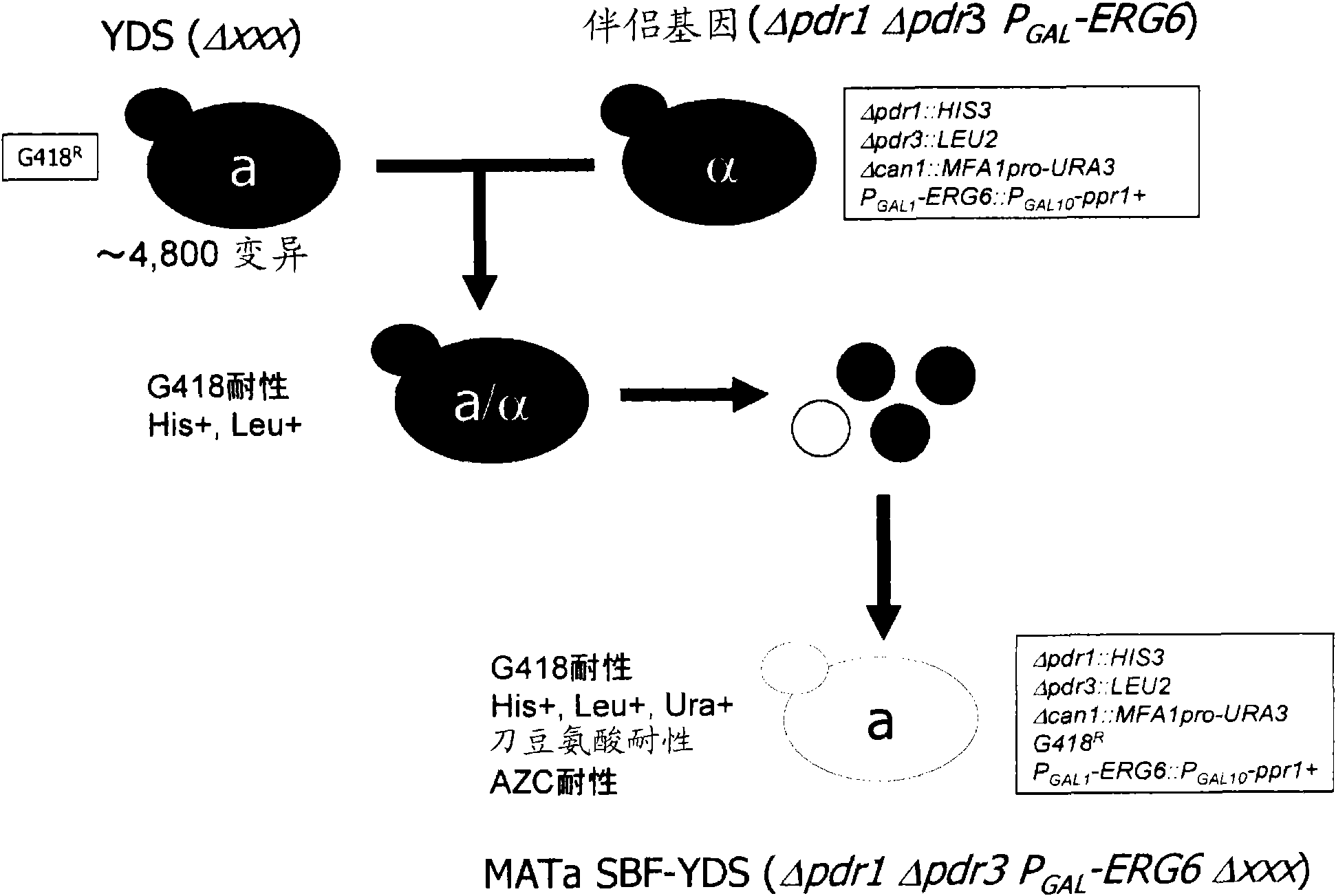
[0079] The first method for identifying a target candidate molecule of a test substance according to the present invention is characterized in that it includes the steps of: contacting the yeast cells contained in the above-mentioned yeast cell set with the test substance; The step of yeast cells with changed phenotype; the step of identifying the mutated gene as the first gene in the above-mentioned selected yeast cells; and the target of using the transcription product and / or translation product of the above-mentioned first gene as a substance to be detected Steps in which candidate molecules are identified.
[0080] The first...
Embodiment 1
[0121]
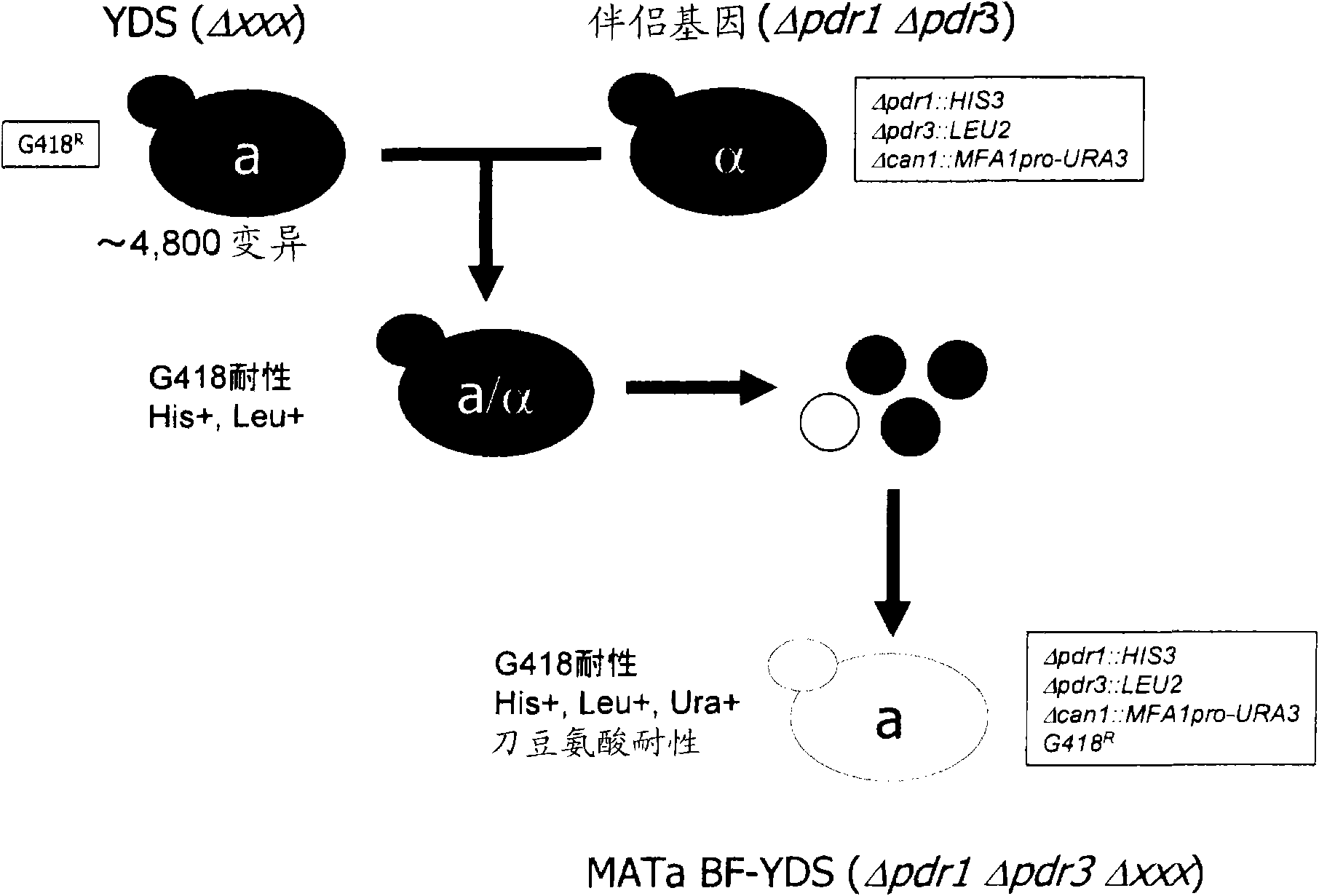
[0122] (1) Preparation of a partner strain in which the PDR1 gene and the PDR3 gene are disrupted
[0123] Using the method described in PCR-based method for producing efficient gene-disrupted alleles (Baudin A. et al. (1993) Nucl. Acid Res. 21: 3329-3330), PDR1 gene and PDR3 gene-disrupted alleles were prepared as follows: Companion strains.
[0124] The PDR3 gene locus of the BY4741 strain (MATa) was replaced with the LEU2 gene, thereby disrupting the PDR3 gene (hereinafter referred to as KE383). Also, the PDR1 gene locus of the BY4742 strain (MATα) was replaced with the HIS3 gene, thereby disrupting the PDR1 gene (hereinafter referred to as KE384). The KE383 strain and the KE384 strain were mated to form spores to produce a Δmet15, MATa-type strain (hereinafter referred to as KE445) in which both PDR1 and PDR3 were disrupted.
[0125] (2) Introduction of the URA3 gene whose expression is induced by the MFA1 promoter into the CAN1 region
[0126] First, using t...
Embodiment 2
[0137]
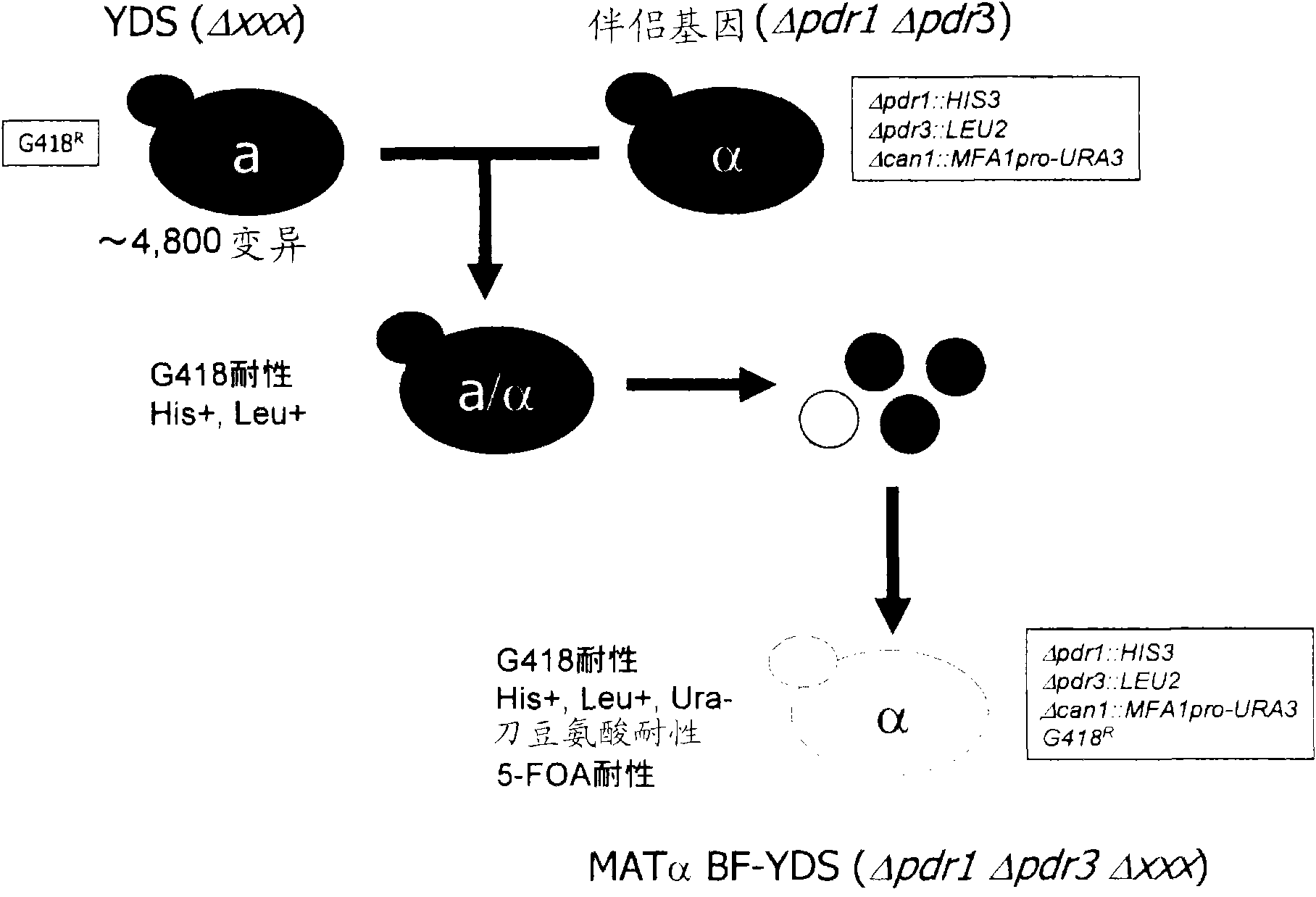
[0138] In Example 1, except that the counter selection using 5-FOA (5-fluoroorotic acid) was carried out instead of selecting yeast cells having the MATa gene, a yeast gene knockout having the MATα gene was prepared in the same manner as in Example 1. Strains set. Specifically, by using MATα selective agar medium (SD-(Met+,Ura+) / G418 / canavanine / 5FOA) instead of MATa selective agar medium (SD-(Met+) / G418 / canavanine), successfully prepared A set of yeast gene knockout strains (MATα type BF-YKO) composed of 1374 strains of yeast cells with MATα gene disrupted by PDR1 gene and PDR3 gene ( figure 2 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com