RNA sequence secondary structure prediction method based on base fragment coding and ant colony optimization
A technology of secondary structure and ant colony algorithm, applied in the field of bioinformatics research, can solve the problems of high cost and difficulty, and achieve the effect of intuitive and accurate structural expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
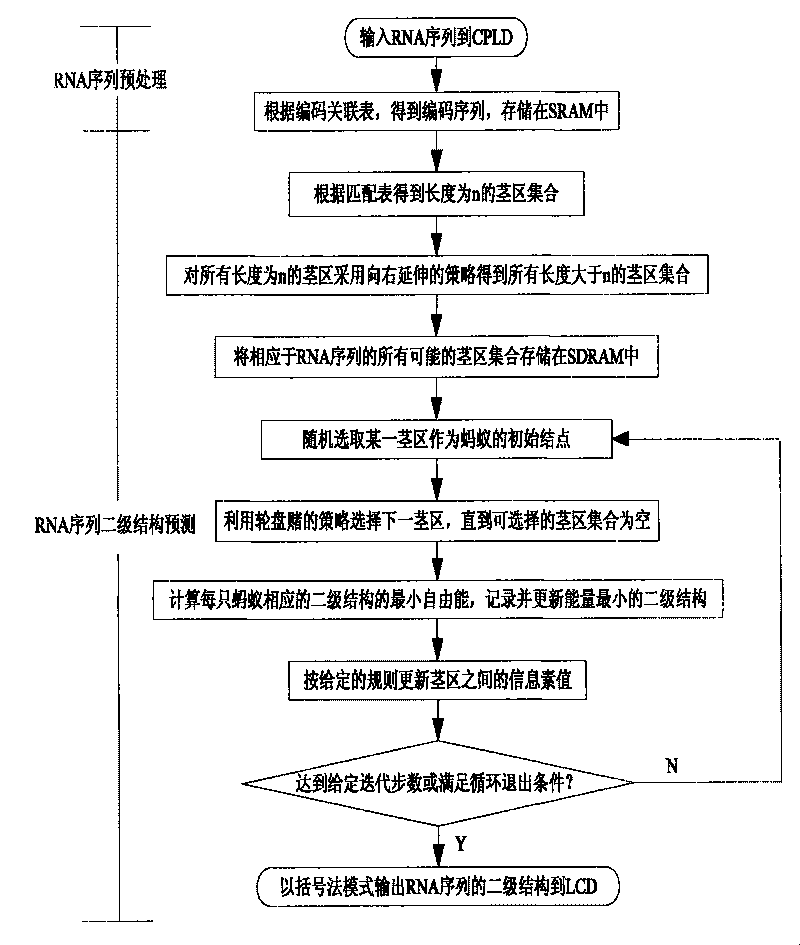
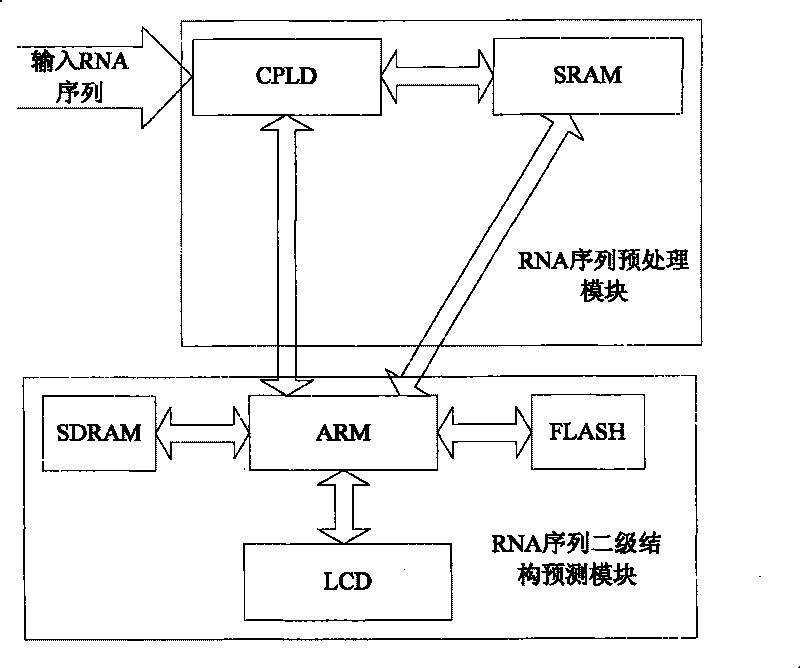
[0021] The present invention is a method for predicting the secondary structure of RNA sequences based on base segment coding and ant colony algorithm, such as figure 1 As shown, the obtained RNA sequence is input into CPLD, and the RNA sequence is encoded through the coding association table, so that the RNA sequence is stored in the SRAM in the form of a coding sequence, and a set of stem regions with a length of n is obtained according to the matching table. The stem region of n adopts the strategy of extending to the right to obtain all the stem region sets whose length is greater than n, and stores all possible stem region sets corresponding to the RNA sequence in SDRAM to be called, and then randomly selects a certain stem region set through the ARM control chip One stem area is used as the initial node of the ant colony algorithm, and the next stem area is selected using the roulette strategy until the selectable stem area set is empty, and finally the minimum free energ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com