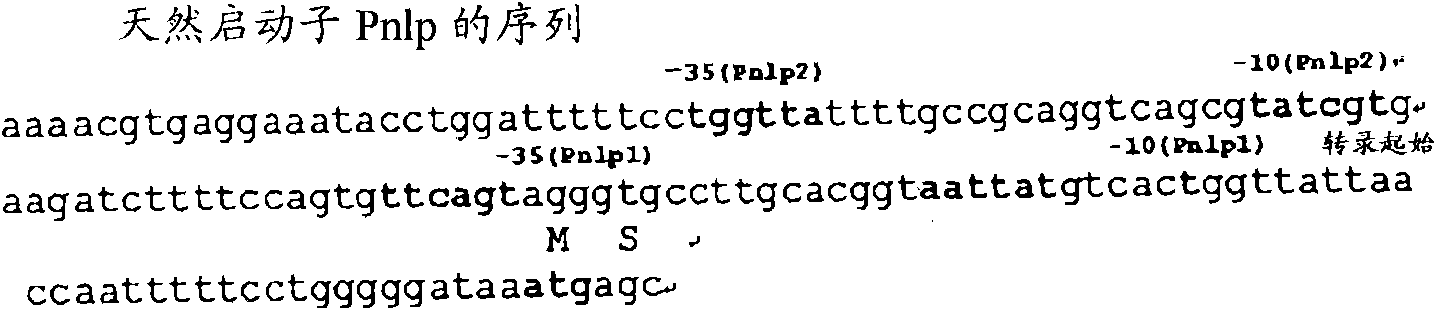An L-amino acid-producing bacterium and a method for producing an L-amino acid
An amino acid and leucine technology, which is applied in microorganism-based methods, biochemical equipment and methods, botanical equipment and methods, etc., can solve the problem of unknown mutation of yeaS gene, and achieve the effect of high-efficiency fermentation production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
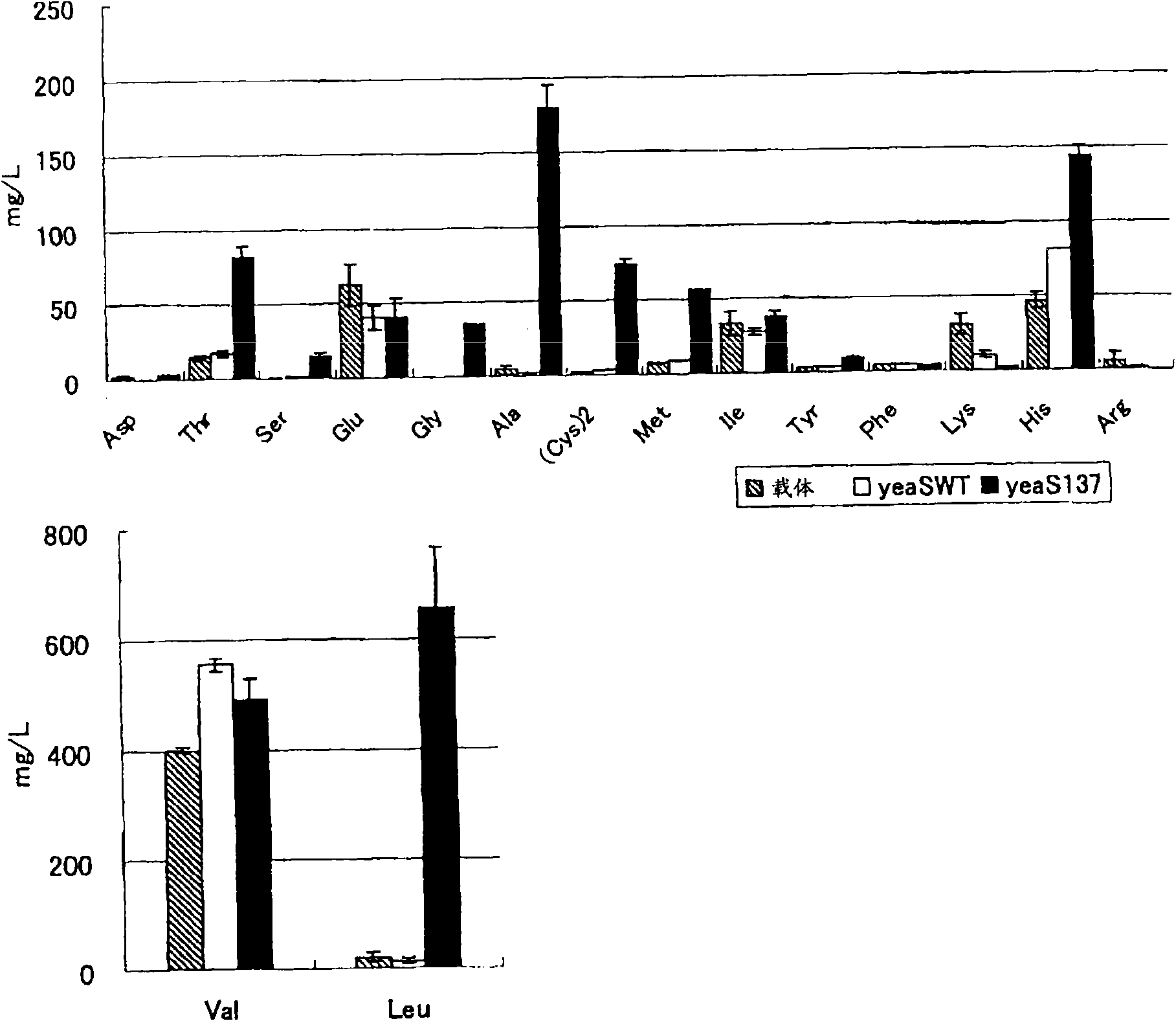
[0242] (Example 1) Effect of increasing L-cysteine production by strengthening wild-type YeaS
[0243] In order to study the effect on L-cysteine production due to enhanced expression of the wild-type yeaS gene in P. ananatis, a gene cysE5 encoding a mutant SAT was introduced (US Patent Application No. 20050112731), and A strain having an increased copy number of the yeaS gene.
[0244] First, the plasmids used to construct the above strains were constructed. Its method is as follows.
[0245] Using the chromosomal DNA of Escherichia coli MG1655 (ATCC No.47076) as a template, using P1 (agctgagtcg acccccagga aaaattggtt aataac: SEQ ID NO: 20) and P2 (agctgagcat gcttccaact gcgctaatga cgc: SEQ ID NO: 21) as primers, obtained by PCR A DNA fragment of about 300 bp containing the promoter region of the nlpD gene (hereinafter, the wild-type nlpD gene promoter is referred to as "Pnlp0") was obtained. Sites for restriction endonucleases SalI and PaeI were designed at the 5' end a...
Embodiment 2
[0259] (Example 2) Obtaining the mutant yeaS gene and L-cysteine production increasing effect
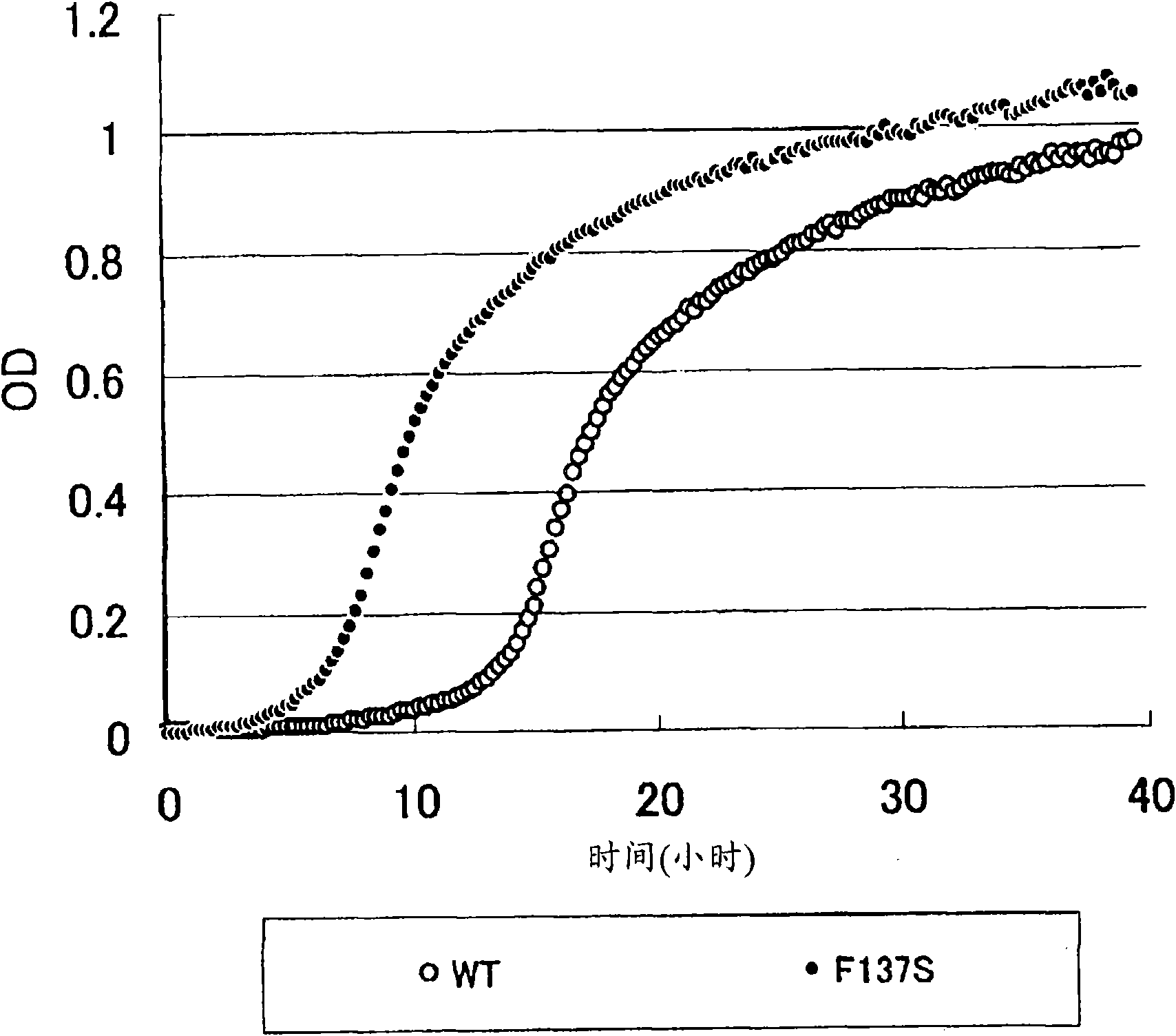
[0260] Next, random mutations were artificially introduced into the yeaS gene by error-prone PCR in order to obtain a mutant with a higher activity of increasing the L-cysteine production capacity of the host cell than the wild-type YeaS.
[0261] (1) Preparation of error-prone PCR and mutation introduction library of yeaS
[0262] First, the error-prone PCR conditions for introducing 1 to 3 mutations into the yeaS gene were based on existing knowledge (Evert Bokma et al., (2006) FEBS Letters 580:5339-5343, Zao et al., (1998) Nat Biotechnol Mar; 16(3):258-61). DNA polymerase uses taq polymerase (manufactured by QIAGEN), and the composition of the reaction solution is 10mM Tris-HCl (pH8.3), 50mM KCl, 7mM MgCl 2 , 0.2mM dGTP, dATP, 1mM dCTP, dTTP and 12.5μM MnCl 2 . The primers used were P9 (catgccatgg tcgctgaata cggggttctg, SEQ ID NO: 28) and P10 (aactgcagtc aggattgcag cgtcgc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com