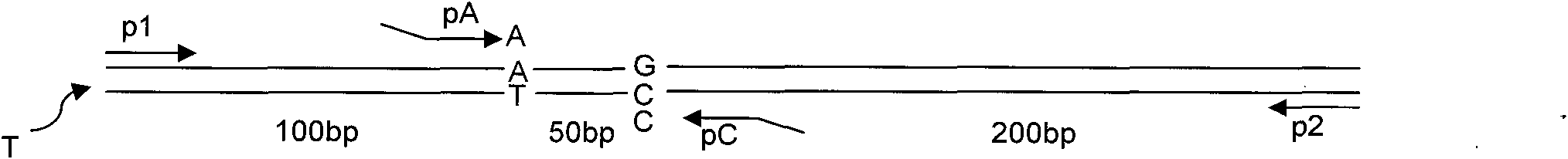New bidirectional one-step genotyping method through PCR
A genotyping and new method technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as high operating costs and complex equipment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
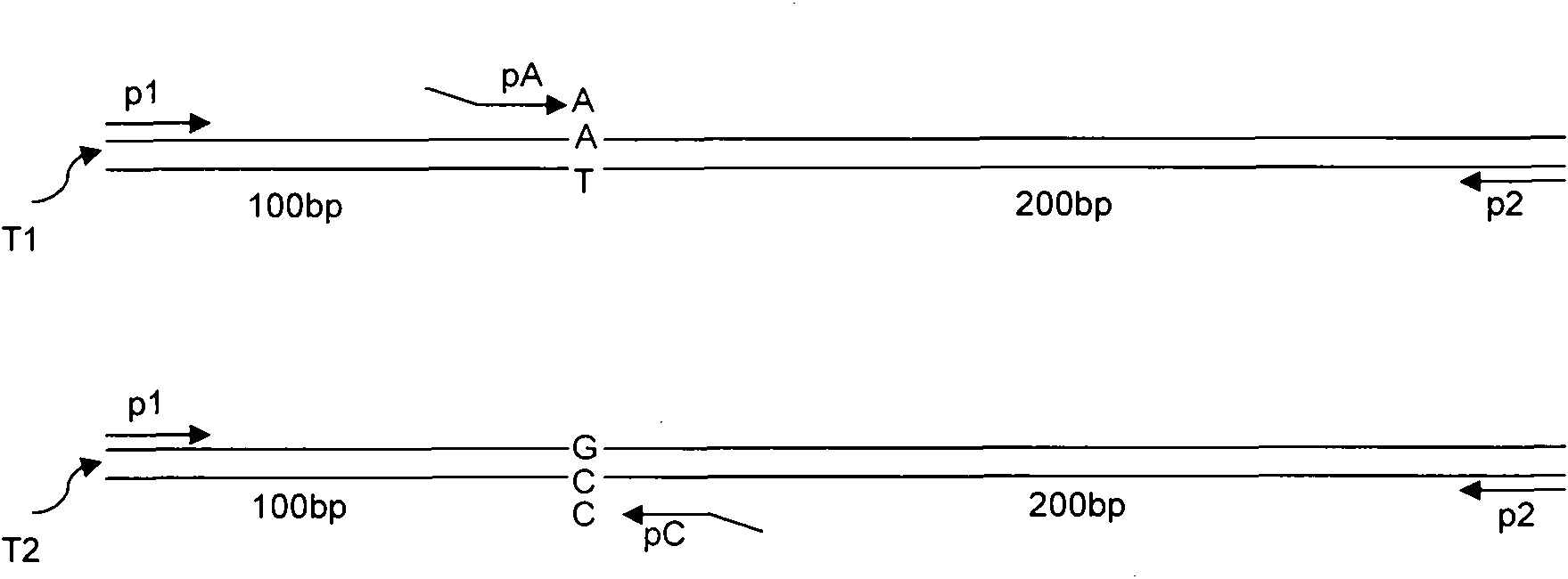
[0039] A two-way one-step PCR genotyping method for the determination of homozygous A / A genotype DNA ( figure 1 T1 in
[0040] Proceed as follows:
[0041] a) According to the genotype A / A of the SNP site, design 5' end template mismatched specific PCR primers pA and pC;
[0042] b) Using specific PCR primers pA and pC that do not match the template at the 5' end, perform PCR from the SNP site in two directions at the same time;
[0043] c) Design two outer-end primers p1 and p2 at different lengths at the far end of the SNP site, whose 5'-3' points to the SNP site from the far end of the SNP site. Used to control the length of PCR products;
[0044] d) Qualitative or quantitative detection by electrophoresis and fluorescent probes.
[0045] The PCR reaction volume is 25 microliters. The pH value of the Tris-HCl buffer system is 8.3. The concentration of Mg ions is 2.5mM, the concentration of dNTP is 200uM, the outer primers are p1 and p2, and the specific primers are pA an...
Embodiment 2
[0048] A two-way one-step PCR genotyping method for the determination of homozygous G / G genotype DNA ( figure 1 T2 in
[0049] Step is with embodiment 1
[0050] The primers are still p1, p2, pA and pC. The PCR reaction volume is 25 microliters. The pH value of the buffer system containing Tris-HCl is 8.3. The concentration of Mg ions is 2.5mM, the concentration of dNTP is 200uM, the concentration of each primer is 200nM, and the KlenTaq DNA polymerase used in the reaction is 1.2u in the total reaction system, 2ng of DNA was used as a template. Negative control samples replaced the DNA template with water.
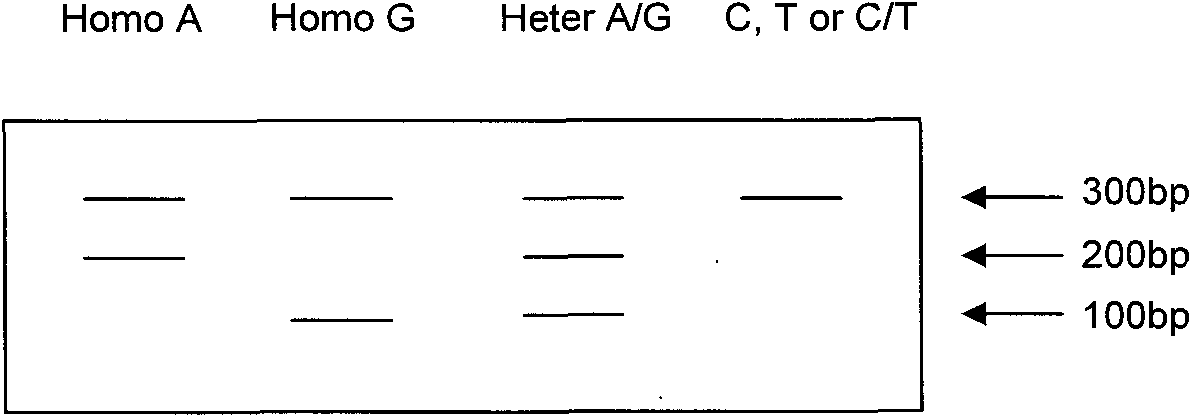
[0051] Electrophoresis results: only a 300bp PCR product synthesized by primers p1 and p2 and a 100bp PCR product synthesized by primers pC and p1. Negative control samples were water free of PCR products.
Embodiment 3
[0053] Two-way one-step PCR genotyping method to determine heterozygous A / G genotype DNA ( figure 1 T1 and T2 in
[0054] Step is with embodiment 1
[0055] The primers are still p1, p2, pA and pC. The PCR reaction volume is 25 microliters. The pH value of the buffer system containing Tris-HCl is 8.3. The concentration of Mg ions is 2.5mM, the concentration of dNTP is 200uM, the concentration of each primer is 200nM, and the KlenTaq DNA polymerase used in the reaction is 1.2u in the total reaction system, 2ng of DNA was used as a template. Negative control samples replaced the DNA template with water.
[0056] Electrophoresis results: There are PCR products synthesized by all primers. That is, a 300bp PCR product synthesized by primers p1 and p2, a 100bp PCR product synthesized by primers pC and p1, and a 200bp PCR product synthesized by primers pA and p2. Negative control samples were water free of PCR products.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com