Method for identifying bifidobactirium
A technology of bifidobacteria and analysis method, which is applied in the field of molecular biology to identify bifidobacteria, and can solve the problems of distinguishing species, difficult bacteria, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] The identification of embodiment 1 Bifidobacterium lactis BL99
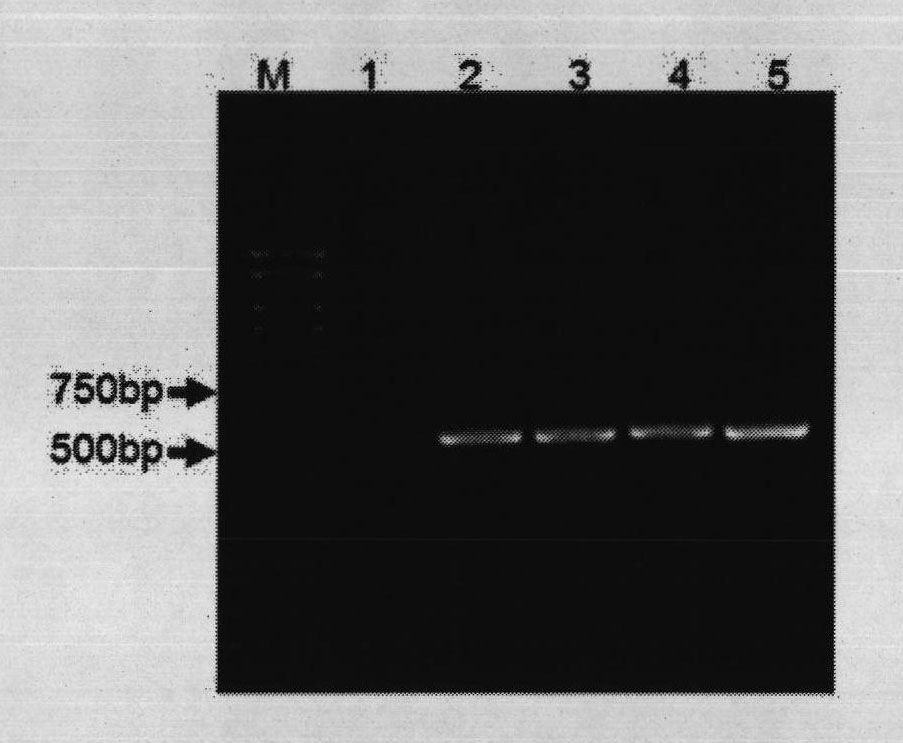
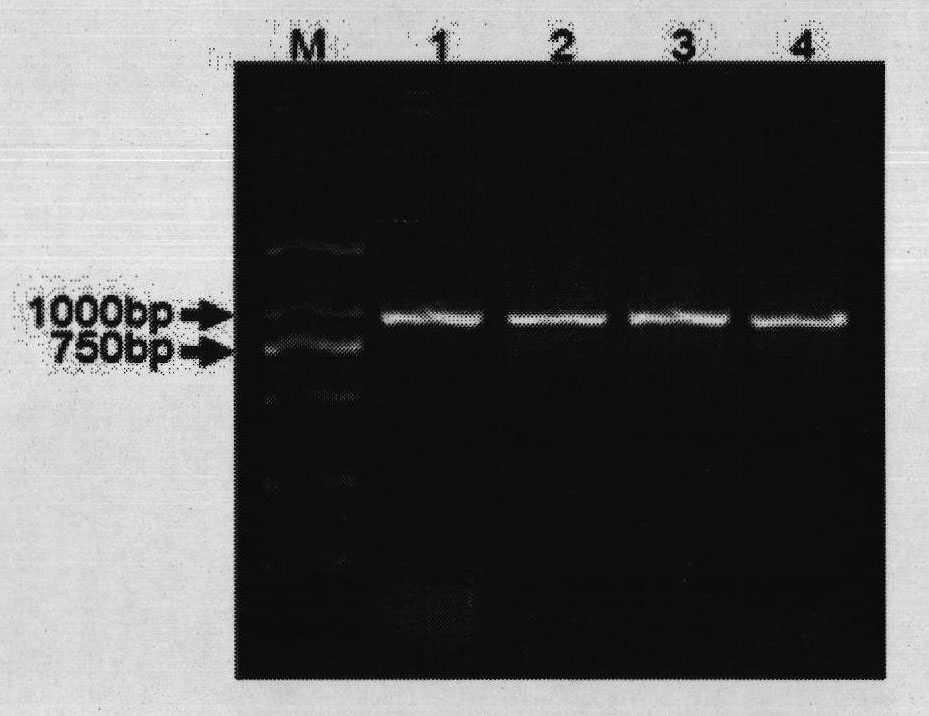
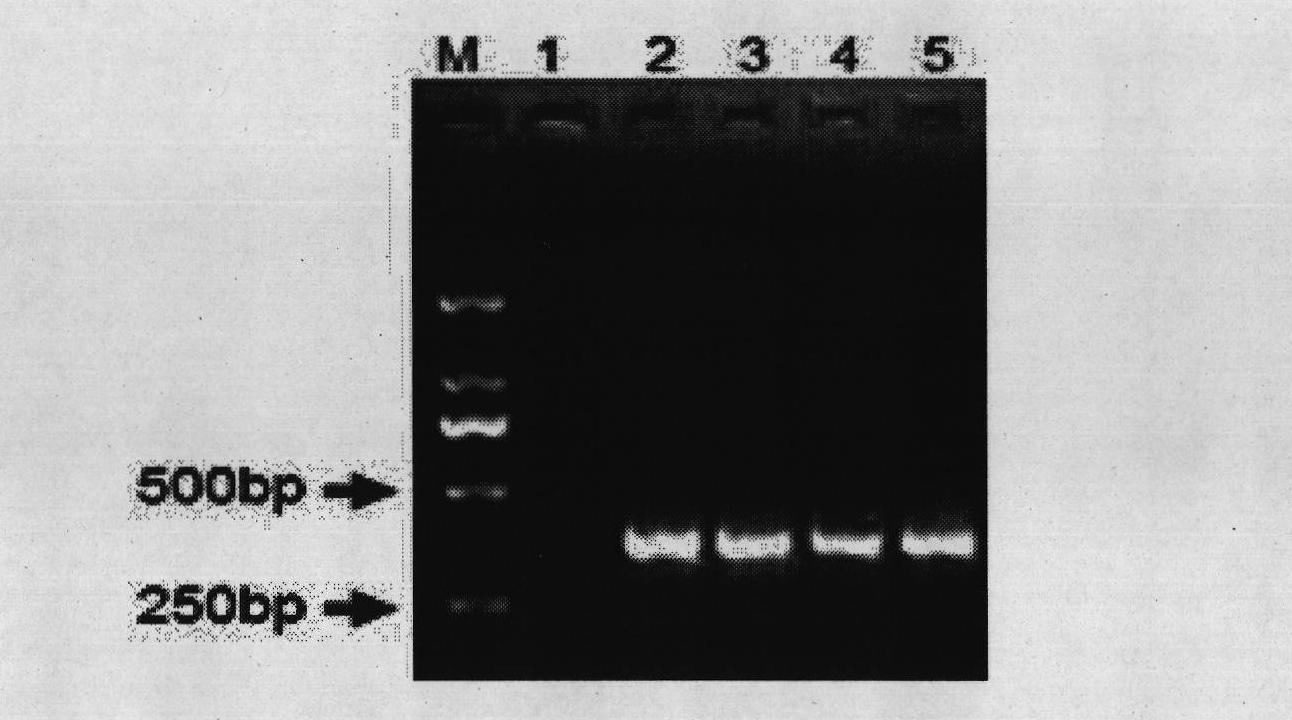
[0061] After the Bifidobacterium lactis strain BL99 was anaerobically cultured in the MRS liquid medium for 24 hours, 1ml of the bacterial liquid was taken to extract genomic DNA, and the DNA of the bacterial strain was used as a template, and the genus-specific primer pair Bif500F & Bif500R was used as primers for PCR amplification. Strain BL99 can be amplified to obtain a target fragment of about 500bp, indicating that the strain belongs to the genus Bifidobacterium; at the same time, primer pairs Bif900F & Bif900R and Pk300F & Pk00R were used for PCR amplification, and the electrophoresis of the amplified product is as follows figure 1 shown.
[0062] The obtained three target gene fragments were purified and sequenced respectively, and the following results were obtained:
[0063] Alignment results of target fragment 1 (Bif500 primer pair): the strains with the highest homology include Bifidobacterium...
Embodiment 2
[0067] Identification of Example 2 Bifidobacterium longum BL88-Onlly
[0068] After the Bifidobacterium longum strain BL88 was anaerobically cultured in the MRS liquid medium for 24 hours, 1ml of the bacterial liquid was taken to extract genomic DNA, and the DNA of the bacteria was used as a template, and the genus-specific primer pair Bif500F & Bif500R was used as primers for PCR amplification. The strain BL88-Onlly can be amplified to obtain a target fragment of about 500bp, indicating that the strain belongs to the genus Bifidobacterium. Carry out PCR amplification with primer pair Bif900F & Bif900R and Pk300F & Pk00R simultaneously, the electrophoresis figure of amplified product is as follows figure 1 shown.
[0069] The obtained three target gene fragments were purified and sequenced respectively, and the following results were obtained:
[0070] Alignment results of target fragment 1 (Bif500 primer pair): the strains with the highest homology include Bifidobacterium l...
Embodiment 3
[0074] Example 3 Identification of Bifidobacterium breve BB8
[0075] After the Bifidobacterium breve strain BB8 was anaerobically cultured in the MRS liquid medium for 24 hours, 1 ml of the bacterial liquid was taken to extract genomic DNA, and the DNA of the bacteria was used as a template, and the genus-specific primer pair Bif500F & Bif500R was used as primers for PCR amplification. Strain BB8 can be amplified to obtain a target fragment of about 500bp, indicating that the strain belongs to the genus Bifidobacterium. Simultaneously carry out PCR amplification with primer pair Bif900F & Bif900R and Pk300F & Pk00R, the electrophoresis figure of amplified product is as follows figure 1 shown.
[0076] The obtained three target gene fragments were purified and sequenced respectively, and the following results were obtained:
[0077] Alignment results of target fragment 1 (Bif500 primer pair): the most homologous strain of Bifidobacterium breve B.breve. The target fragment s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com