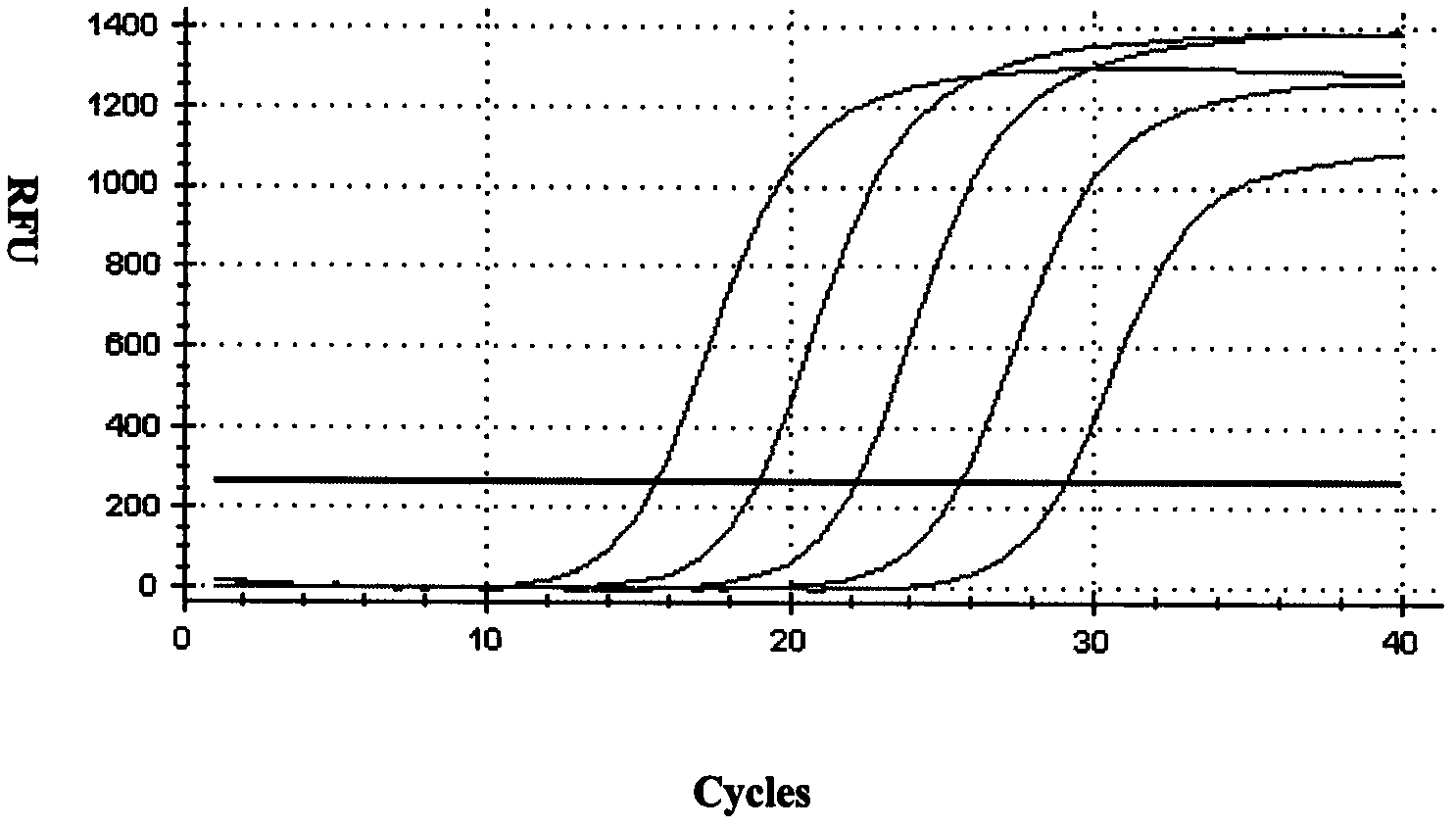
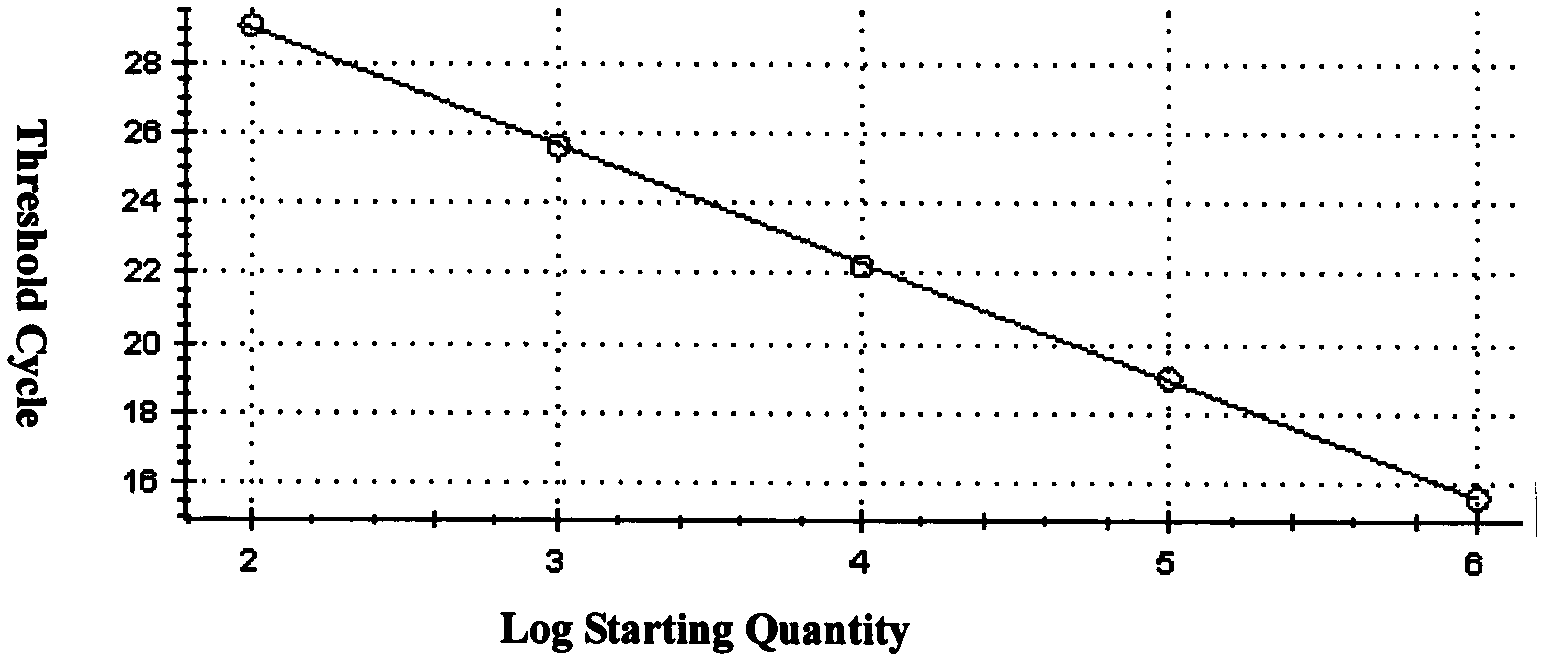
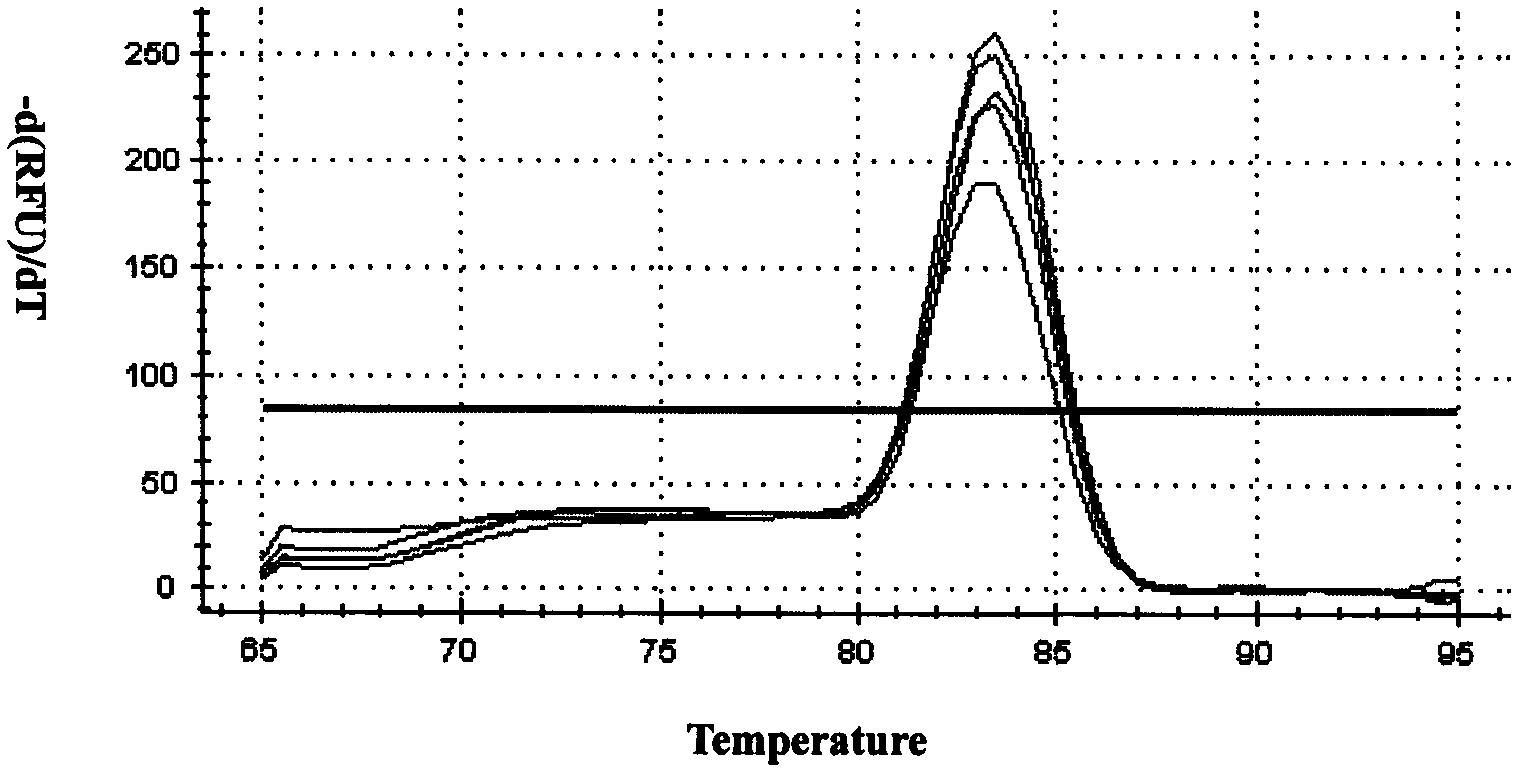
Method for screening real-time fluorescent quantitative PCR internal reference molecules of desert plant Eremosparton songoricum (Litv.) Vass.
A real-time fluorescence quantitative and internal reference technology, applied in biochemical equipment and methods, fluorescence/phosphorescence, microbial measurement/inspection, etc., can solve the problems of blank research on internal reference molecules and unreported internal reference gene sequence data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0125] In the following examples, the experimental methods without specific conditions are usually carried out according to routine experimental operations. It was carried out according to the method described in "Refined Molecular Biology Experiment Guide" (Edited by F.M Osper, R.E. Kingston, J.G Seidman, translated by Ma Xuejun, Shu Yuelong, Beijing: Science Press, 2004).
[0126] Cloning of internal reference molecules:
[0127] Using the published GAPDH, Actin, 18SrRNA, UBQ, EF, α-tubulin, β-tubulin gene sequences in the NCBI database as reference, design specific primers for each internal reference gene, and Two-week-old leafless bean seedlings cultured in an incubator (temperature 25°C, time 12h / 12h) were used as materials, RNA was extracted, cDNA was reversed, and target genes were amplified with designed primers for each internal reference gene, and each amplified gene was recovered. A specific fragment of the gene is added, connected to the cloning vector, sequenced ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com