Bispecific egfr/igfir binding molecules
An IGF-IR, binding technology, applied in the direction of peptide/protein components, growth factors/inducible factors, anti-receptors/cell surface antigens/cell surface determinant immunoglobulins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0361] Example 1: Western Assay in Cells for Screening for −+EGFR Activity
[0362] Developed an In Cell Western assay to screen various 10 The ability of Fn3 monoclonals to inhibit EGFR activity, thus identifying the ability to interact with IGF1R 10Fn3 binders ligated to construct E / I binders. In-cell Western assays were also used to screen and measure specific E / I 10 Relative potency of Fn3 binders. Two in-cell Western assays were developed to measure 1) inhibition of EGF-stimulated EGFR phosphorylation or 2) inhibition of EGF-stimulated ERK phosphorylation. For A431 epidermoid carcinoma or FaDu head & neck carcinoma cells, cells were seeded at 24,000 cells / well into poly-D-lysine-coated 96-well microtiter plates (Becton Dickinson, Franklin Lakes, NJ) medium and allow them to attach overnight. Cells were washed once and then incubated in serum-free medium for 24 hours. Next, based on 10 Serial dilutions of conjugates of Fn3 were applied to cells and incubated for 2-3...
Embodiment 2
[0364] Example 2: Based on 10 Expression of Fn3 binders
[0365] Covalently link EGFR binding using a glycine-serine linker 10 Fn3 binding to IGFIR 10 Fn3, generated from 10 Fn3 dimers, each of which 10 The Fn3 domain binds different targets, resulting in E / I binders. IGFIR binding 10 Fn3(I1) was previously described as SEQ ID NO: 226 in PCT Publication No. WO 2008 / 066752. Two novel EGFR binding agents were identified by screening an RNA-protein fusion library for binders of EGFR-Fc (R&D Systems, Minneapolis, MN) as described in PCT Publication No. WO 2008 / 06675 10 Fn3 (E2 and El). The following examples describe the use of various His-tagged 10 E / I binders of Fn3 (non-PEGylated): E2-GS10-I1 (SEQ ID NO:25), E1-GS10-I1 (SEQ ID NO:31), I1-GS10-E1 (SEQ ID NO: 28), and the results obtained with I1-GS10-E2 (SEQ ID NO: 22).
[0366] The following examples also describe the use of the following PEGylated, His-tagged 10 Results obtained for E / I conjugates of Fn3: E1-GS10-I1...
Embodiment 3
[0373] Embodiment 3: based on 10 PEGylation of E / I binders of Fn3
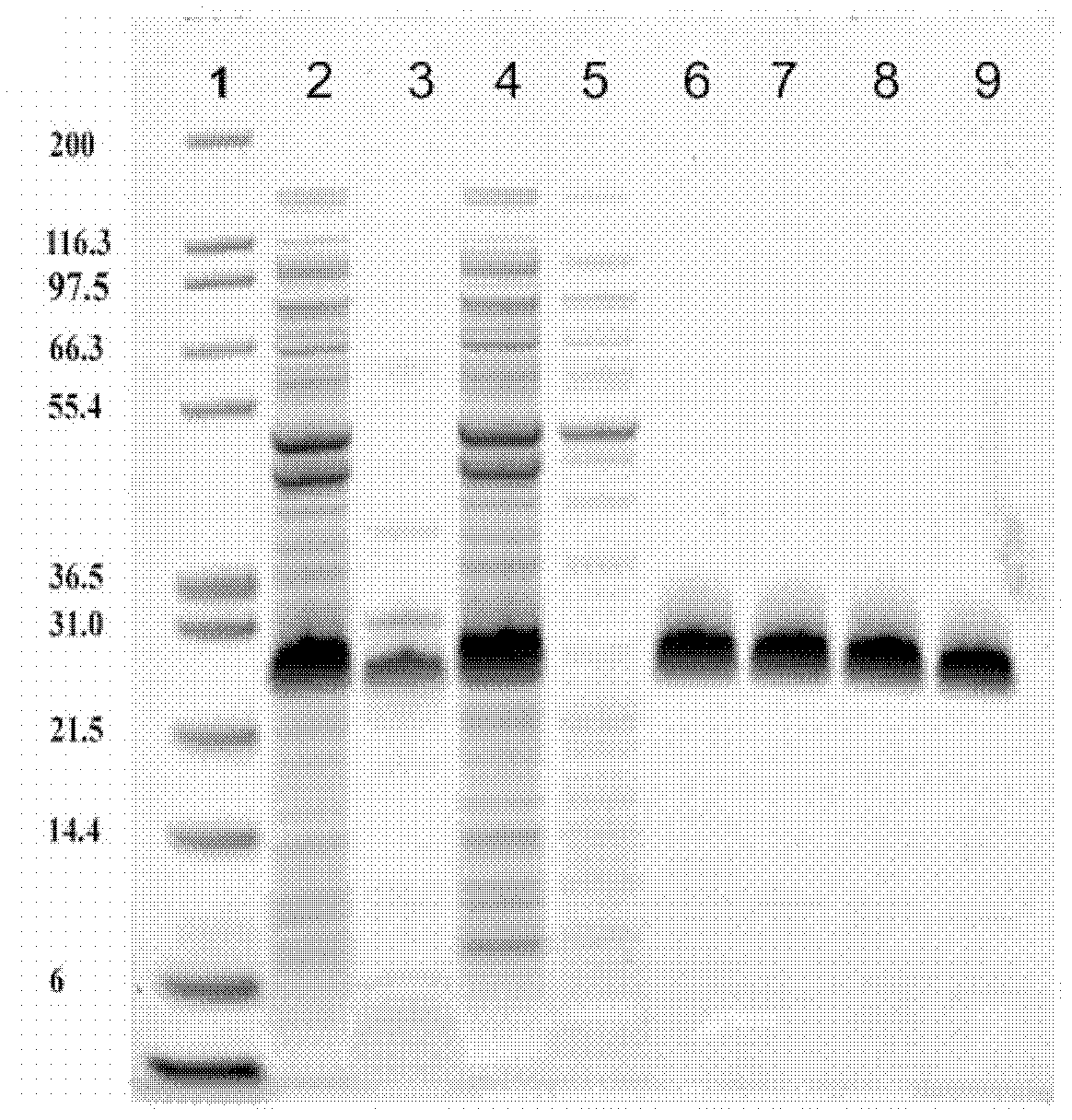
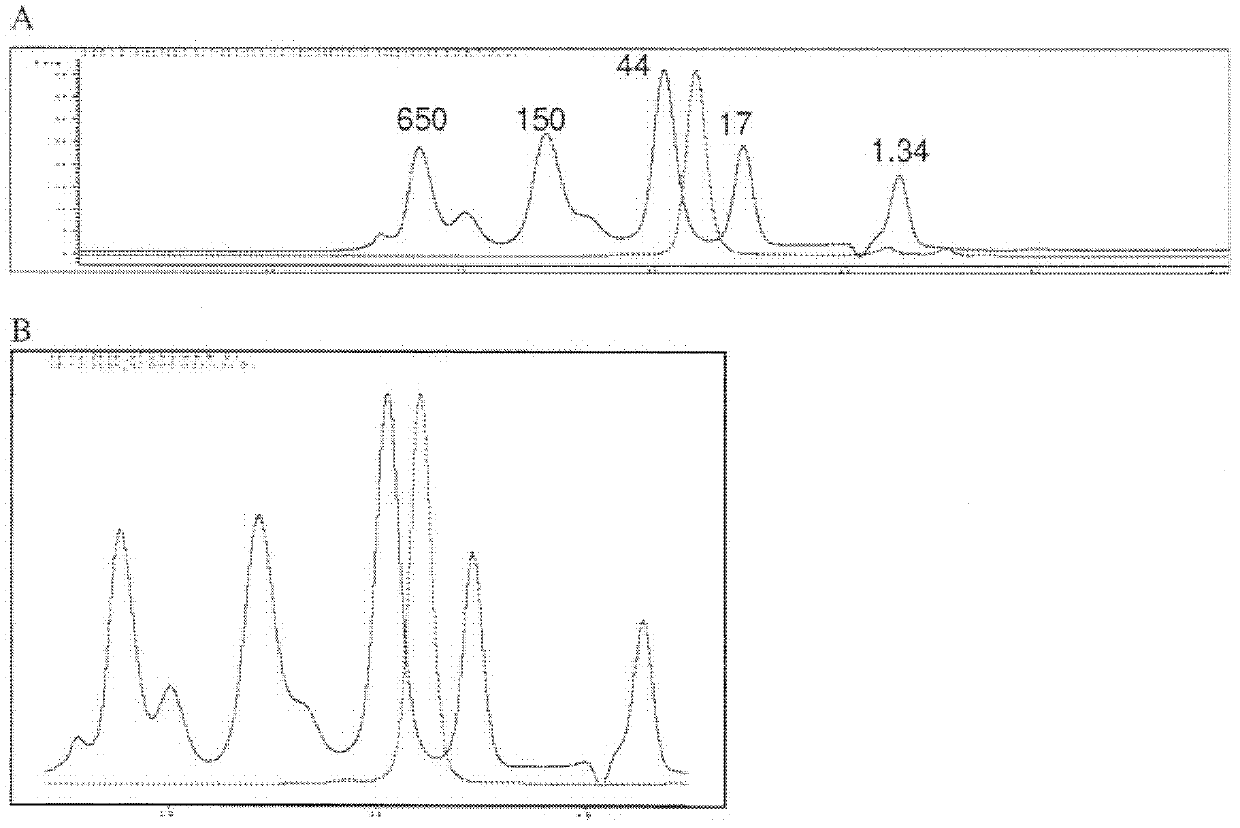
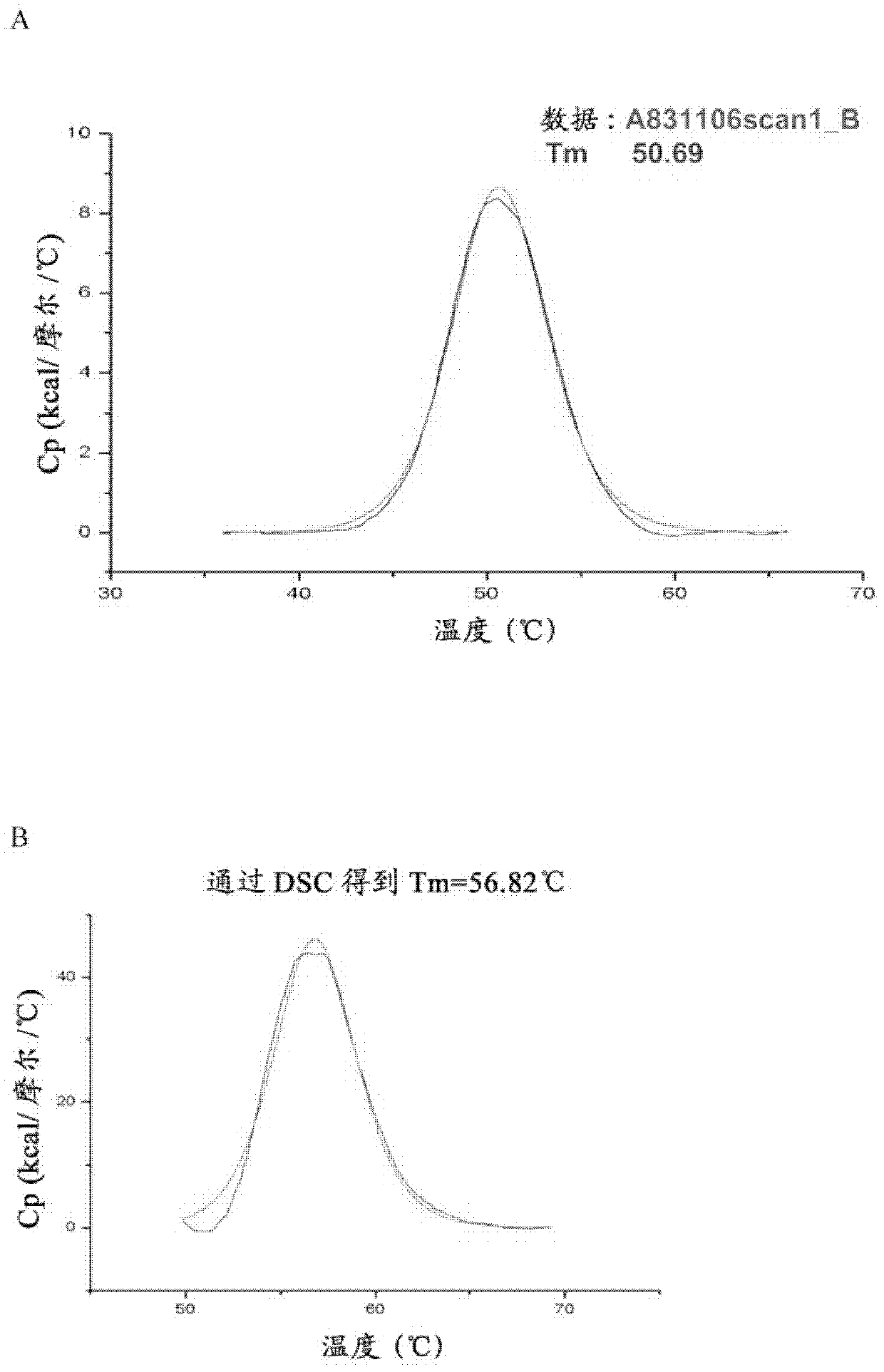
[0374] Fibronectin-based multivalent scaffold proteins (such as those based on 10 E / I binder of Fn3) PEGylation. To allow PEGylation, the modification is usually made near the C-terminus of the protein by single point mutation of an amino acid (usually serine) to cysteine. PEGylation of proteins at single cysteine residues is achieved by mixing and incubating derivatized PEG reagents with protein solutions to conjugate various maleimide-derivatized PEG forms. Proceeding and confirmation of the PEGylation coupling reaction can be confirmed by SDS-PAGE and / or SE-HPLC to separate the non-PEGylated protein from the PEGylated protein.
[0375] For example, construct E2-GS10-I1 (SEQ ID NO: 25) was PEGylated by replacing serine at position 221 with cysteine. The resulting construct, SEQ ID NO: 56, was then coupled to a maleimide derivatized 40 kDa branched PEG (NOF America Corporation, White Plains, NY). The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com