Method for performing sequencing and cluster analysis on V6 hypervariable region of metagenomic 16S rDNA
A technique of metagenomic and clustering analysis, which is applied in the field of sequencing and clustering analysis of metagenomic 16S hypervariable region V6, can solve the problems of insufficient sequencing information, microbial classification, short read length, and inability to sequence 16S rDNA, and saves economic costs. , the effect of reducing human labor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
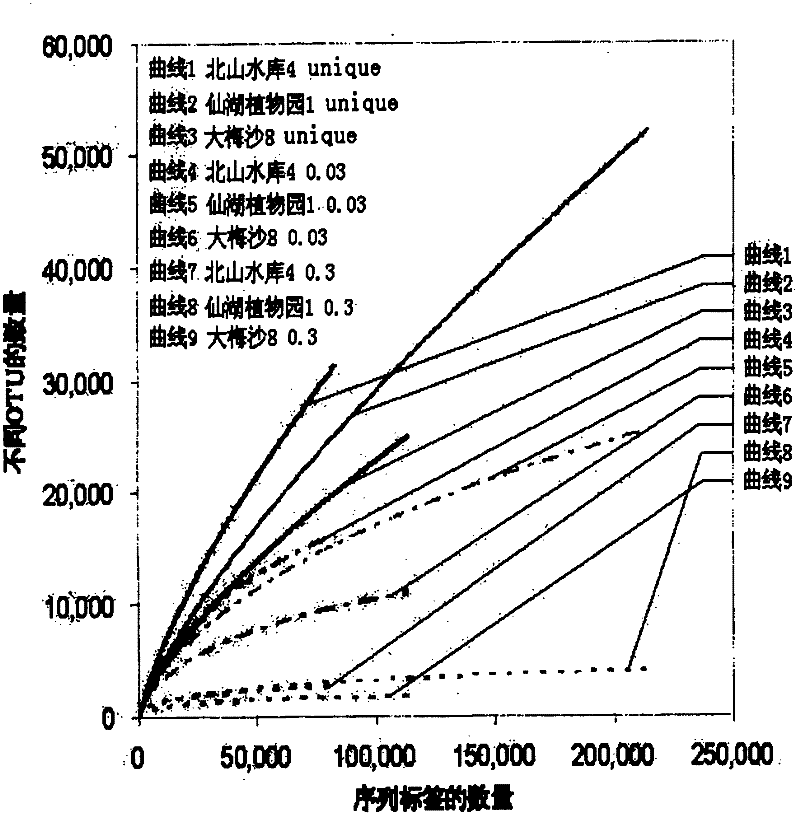
Image
Examples
Embodiment Construction
[0022] The present invention will be described more fully hereinafter with reference to the accompanying drawings, in which exemplary embodiments of the invention are illustrated.
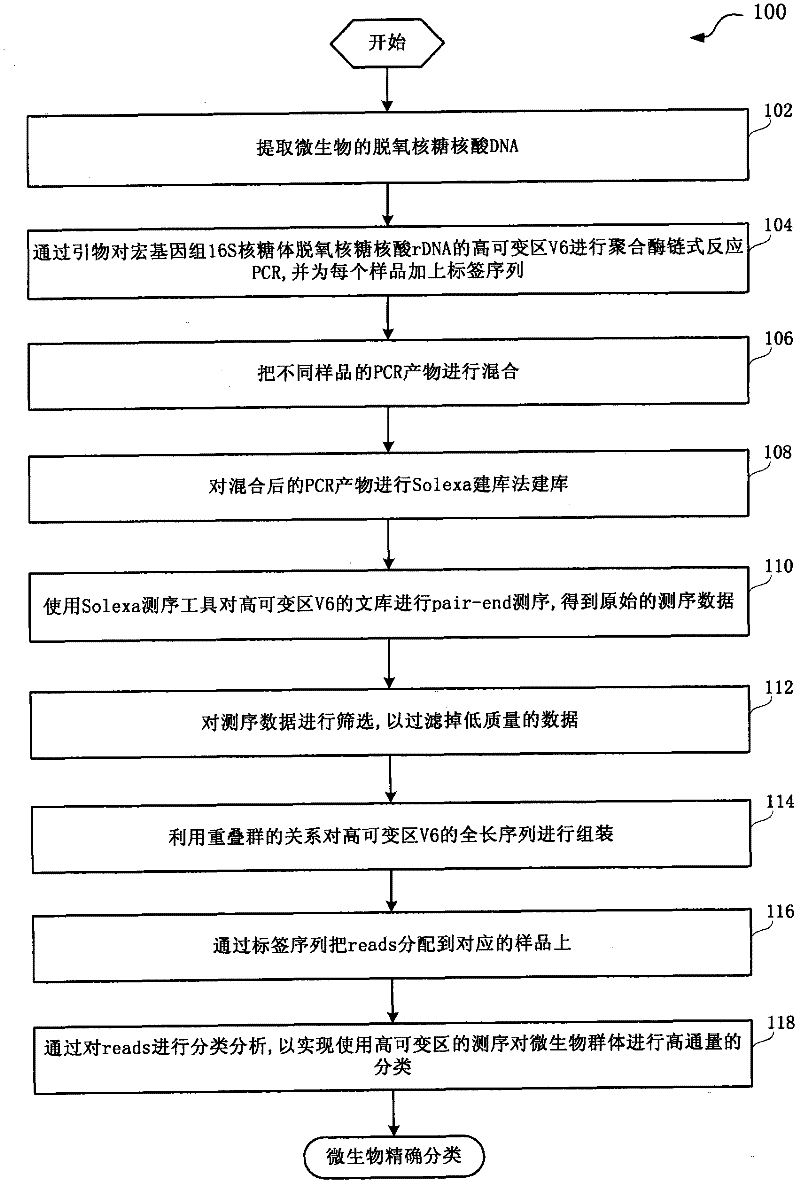
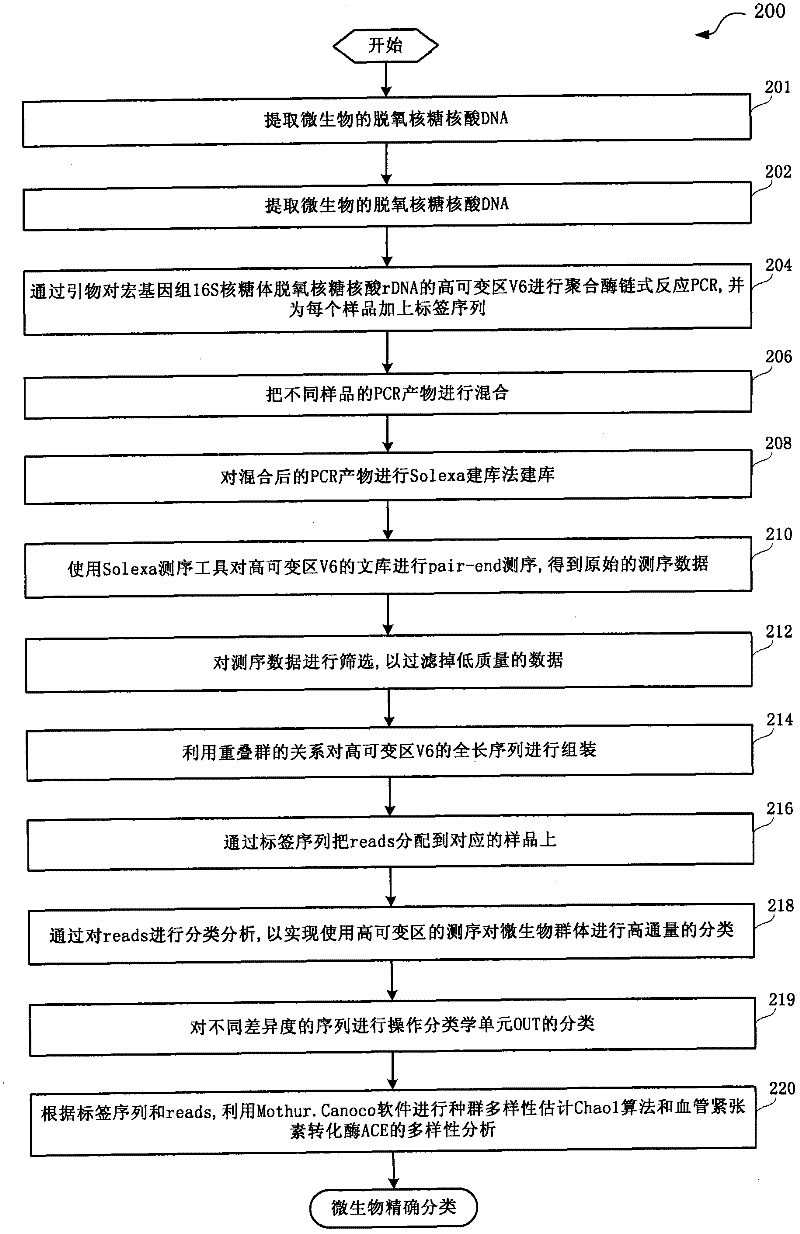
[0023] figure 1 A flow chart showing a method for performing sequencing cluster analysis on the 16S hypervariable region V6 of the metagenomics provided by an embodiment of the present invention.
[0024] Such as figure 1 As shown, the method flow 100 for sequencing and clustering analysis of the metagenomic 16S hypervariable region V6 includes:
[0025] Step 102, extracting the deoxyribonucleic acid DNA of the microorganism. For example, the Ultraclean Soil DNA kit (MoBio, USA) is used to extract microbial DNA from the sample sediment.
[0026] Step 104, the hypervariable region V6 of the metagenomic 16S ribosomal deoxyribonucleic acid rDNA by primers (the two ends of this region each have a conserved region of about 20 base pairs bp, and the variable region in the middle is about 60-90bp) Per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com