Nematode-resistant transgenic plants
A technology of transgenic plants and transcription factors, applied in the field of nematode-resistant transgenic plants, can solve the problem of not deregulation of genetically modified plants, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Embodiment 1: vector construction
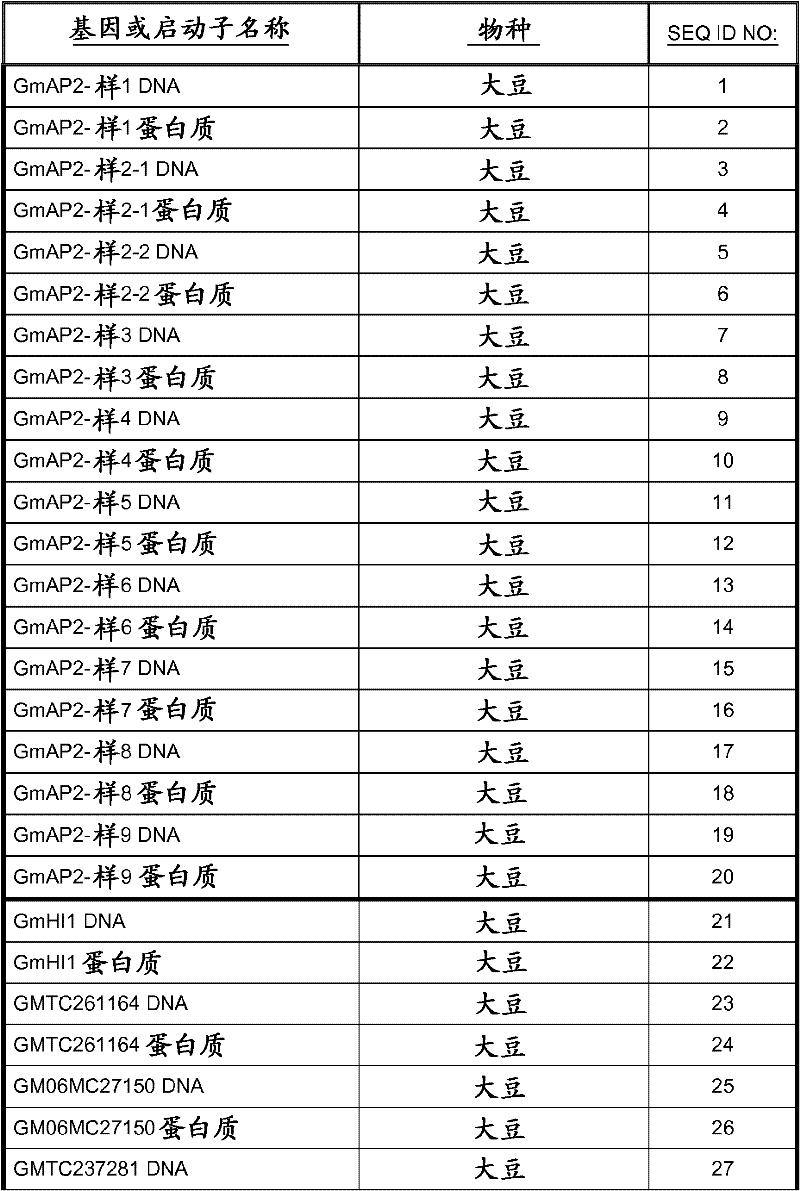
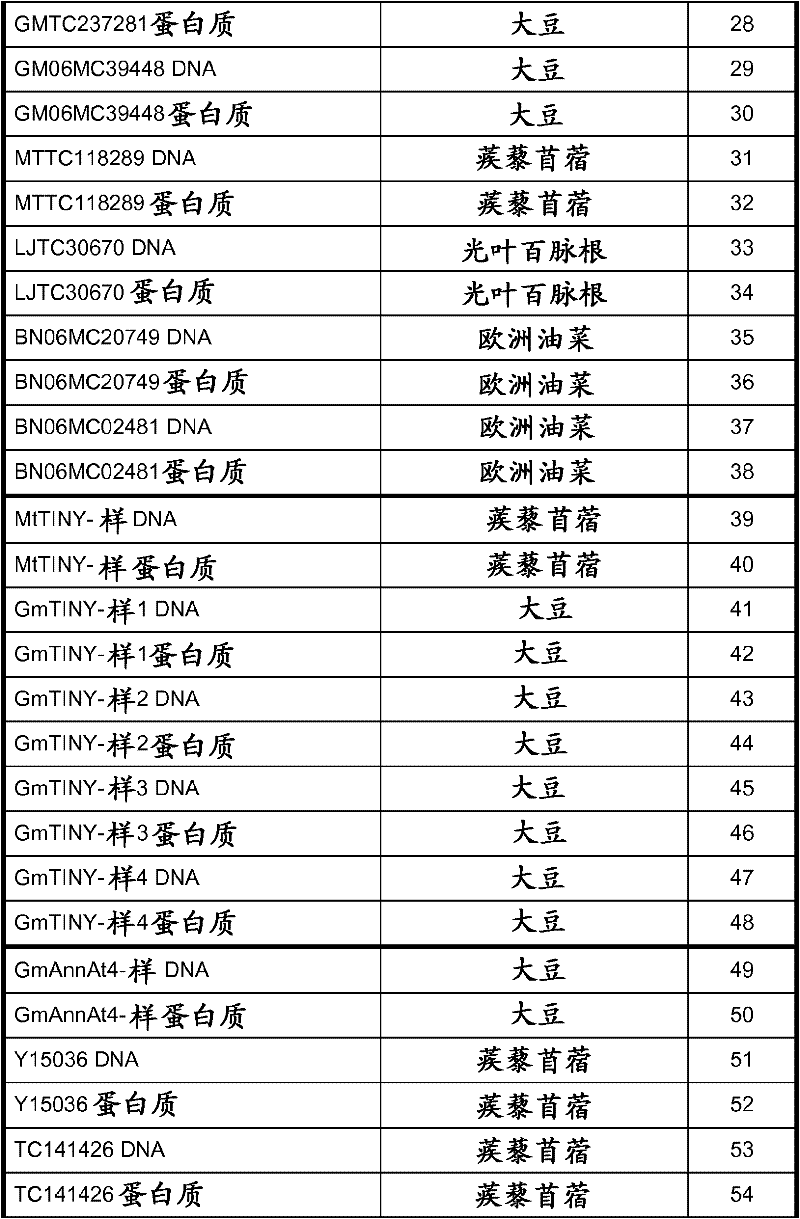
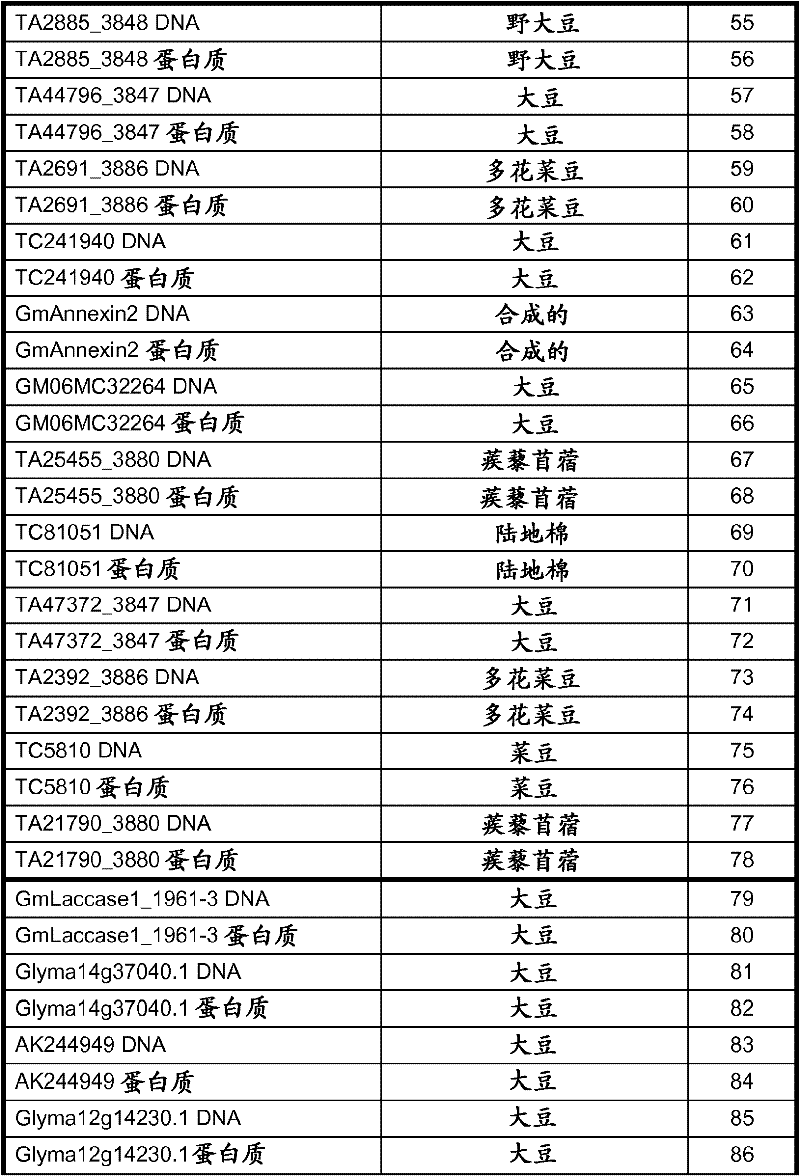
[0080]Using the available cDNA sequences of soybean target polynucleotides, PCR was used to isolate DNA fragments that were used to construct the binary vectors described in Table 1 and discussed in Example 2. The PCR product was cloned into TOPO pCR2.1 vector (Invitrogen, Carlsbad, CA) and the insert was confirmed by sequencing. Using this method to isolate polynucleotides GmAnnAt4-like (SEQ ID NO: 49), GmAux28 (SEQ ID NO: 127), GmIsoflavone7OMT-9 (SEQ ID NO: 117), GmAnUGT_47218626 (SEQ ID NO: 109), Gmhsr201- The open reading frames described by MtTINY-like (SEQ ID NO: 105), MtTINY-like (SEQ ID NO: 39), GmLaccase1 (SEQ ID NO: 79) and GmHI1 (SEQ ID NO: 21). Alternatively, available soybean genomic sequences were used to design primers for amplification from the soybean genomic DNA gene sequence to construct the binary vectors described in Table 1 and discussed in Examples 2 and 3. The DNA sequence of the soybean target gene was PCR ...
Embodiment 2
[0085] Example 2: Nematode Bioassays
[0086] Bioassays to assess nematode resistance conferred by polynucleotides described herein were performed using the rooting plant assay system disclosed in co-owned co-pending USSN 12 / 001,234. Transgenic roots were generated after transformation with the binary vector described in Example 1. Multiple transgenic root lines were subcultured and inoculated with surface-cleared race3 SCN second stage larvae (J2) at a level of approximately 500 J2 / well. Four weeks after nematode inoculation, the number of cysts in each well was counted. For each transformation construct, the number of cysts for each line was calculated to determine the average cyst number and standard error for the construct. The cyst counts for each transformation construct were compared to the cyst counts for the empty vector control tested in parallel to determine whether the tested construct resulted in a reduction in cyst number. Rooted explant cultures transformed w...
Embodiment 3
[0088] Example 3: Root Biomass Determination
[0089] The rooting plant assay system disclosed in co-owned co-pending USSN 12 / 001,234 was also used to assess root growth of uninfected transgenic roots comprising RTP2833, RTP2834, and RTP2839. Multiple transgenic roots and attached cotyledons were subcultured on agar plates for observation. Mark the root tip on the back of the plate as a reference point during subculture. Incubate subcultured roots and cotyledons side by side in the light chamber for up to 6 days. Root weight, root length and number of lateral roots were recorded for each transformation construct. Root parameter measures for each transformation construct were compared to those of the empty vector control tested in parallel to determine whether the tested construct resulted in changes in root weight, root length, root area, and lateral root number. Rooted explant cultures transformed with vectors RTP2833 and RTP2839 showed a general trend of increased root we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com