Method for extracting total DNA (Deoxyribonucleic Acid) of microorganisms from pile-fermentation Pu-erh tea
A compost fermentation and microbial technology, applied in the direction of DNA preparation, recombinant DNA technology, etc., can solve the problems of uncultivable microorganisms and achieve the effect of short experiment time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
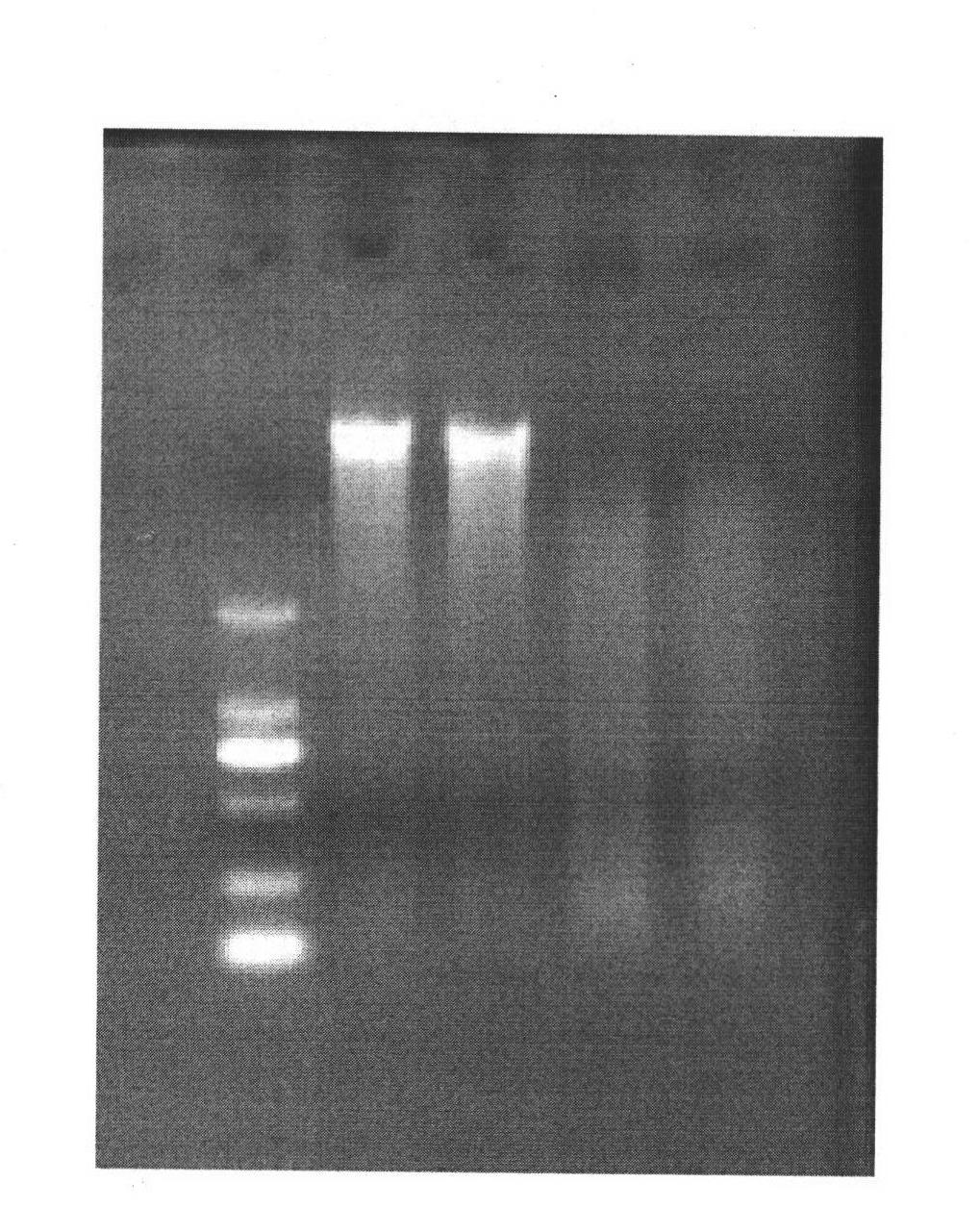
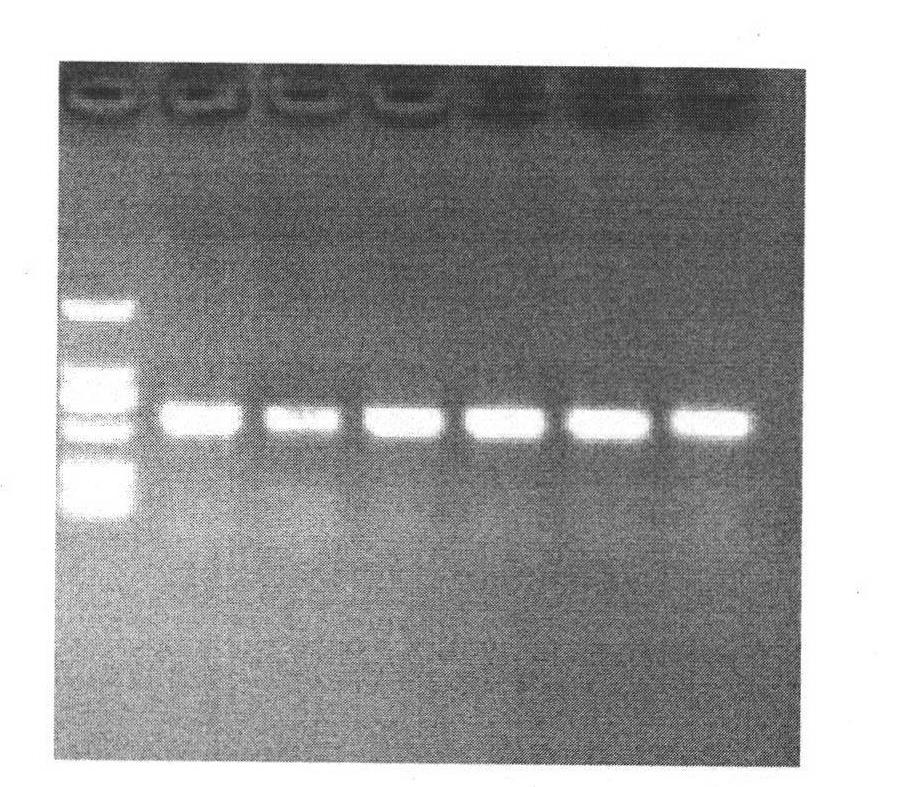
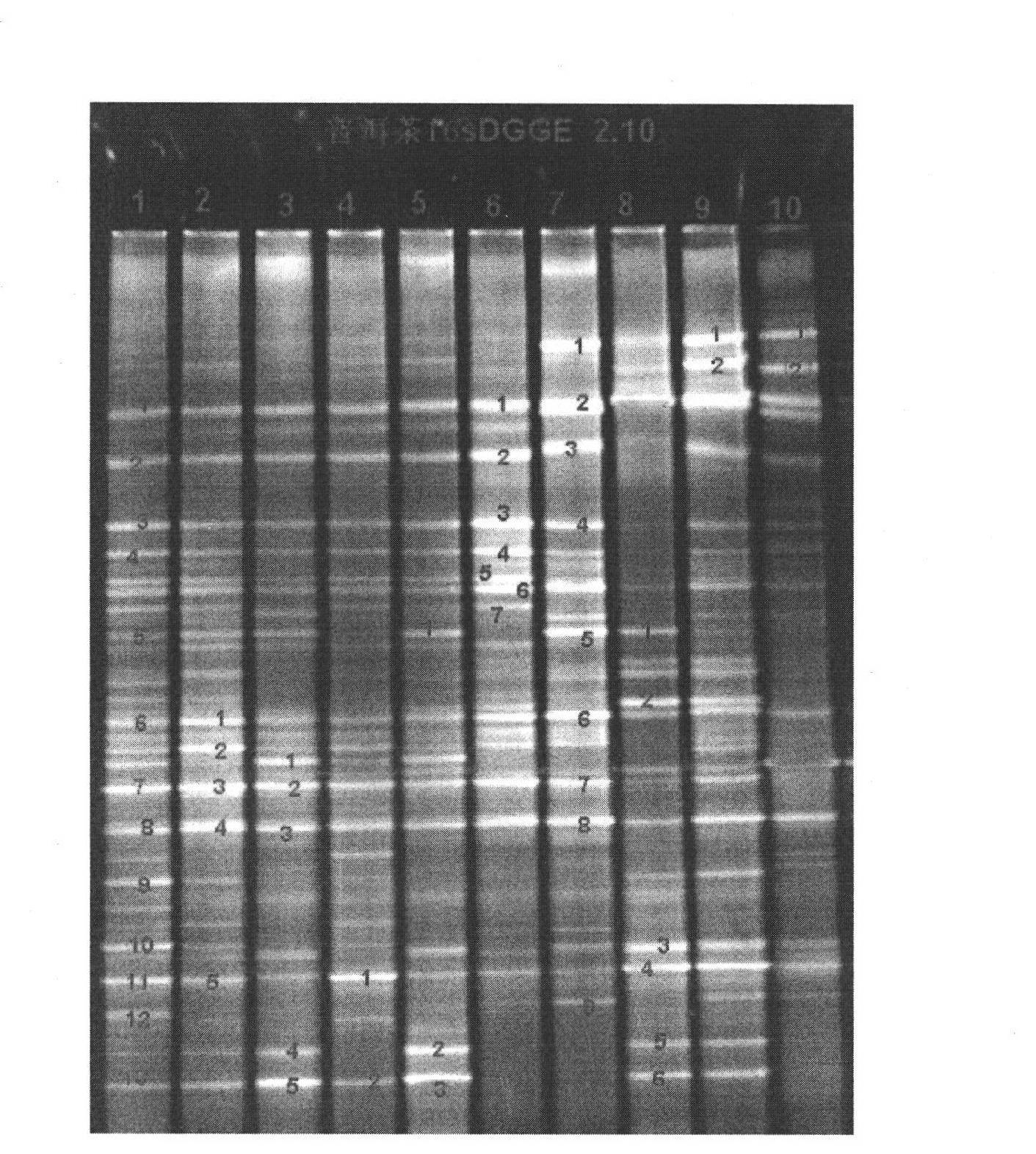
[0032] Sampling of pile-fermented Pu-erh tea, taking pile-fermented Pu-erh tea with different turning times and different stacking positions, analyzing the population structure of bacteria and determining the dominant flora are as follows
[0033] 1) Take 5g tea sample, use liquid nitrogen to freeze and grind 3 times, transfer to a 2ml centrifuge tube, add 200mg glass beads. Use the Omega soil DNA extraction kit to extract the treated tea samples, add 600ul SLX solution, mix evenly with high-speed shaking for 10 minutes, and bathe in 70°C water for 10 minutes. During this period, mix the samples several times, add 200ul SP2 solution, mix well, and ice bath for 5 minutes. Centrifuge at 13000g for 5min. Transfer the supernatant to a new centrifuge tube, add 0.7 times isopropanol, invert and mix well, centrifuge at 13000g for 10min, discard the supernatant, invert on absorbent paper to dry, add 200ul elution buffer to the centrifuge tube, mix well, Dissolve DNA in a water bath a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com