Polypeptide for detecting activity of ATG4B (Autophagy-related protein 4B) in living cell and application thereof
A fusion peptide and reaction buffer technology, applied in the design of new small peptides, the application of cell autophagy level, and the synthesis field, can solve the problems of uncontrollable quality and complicated operation, and achieve strong objectivity, simple operation, and objective reflected effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] 1. Design of peptides
[0056] Based on the substrate sequence TFG of ATG4B, a membrane-penetrating peptide RKKRRQRRR and amino acid residue G are directly coupled at its N-terminus by a peptide bond to form a new type of polypeptide (fusion peptide) (synthesized by Shanghai Gil Biochemical Co., Ltd.) . The amino acid sequence of the polypeptide is RKKRRQRRRGTFG, and the molecular weight is 1701.02.
Embodiment 2
[0058] 2. Design and synthesis of peptide substrates
[0059] Label the N-terminal and C-terminal markers of the polypeptide to obtain a polypeptide substrate. The marker is a small molecular compound for detection, such as fluorescein or nitroaniline salt (pNA), and the marker in this embodiment uses fluorescein. The fluorescent colors of the N-terminal and C-terminal of the labeled polypeptide are different. The N-terminal fluorescence is used as a background reference signal, and the C-terminal fluorescence is used as a detection signal. It is also possible to label the C-terminus of the fusion peptide with fluorescein alone as the detection signal, and use the cell number or total protein amount as the background reference signal. The N-terminus of the polypeptide in this example is labeled with FITC green fluorescence, and the C-terminus is labeled with AMC blue fluorescence. Of course, the N-terminal and C-terminal of the polypeptide can also be labeled with other meth...
Embodiment 3
[0068] 3. Peptides can detect ATG4B activity in cell lysates
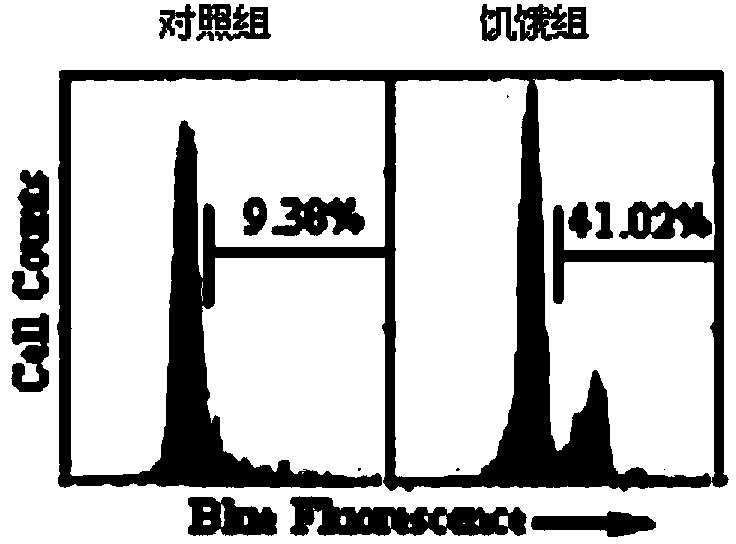
[0069] 1. Autophagy induction model
[0070] In order to detect whether the polypeptide of the present invention has the ability to detect ATG4B activity, a cell autophagy induction model was first constructed. Human cervical cancer cells Hela were treated with 1×10 5 Inoculated into 6-well cell culture plates at a ratio of 2 ml per well. After the overnight culture, the culture medium was removed, the control group was still cultured with normal medium, and the cells in the experimental group were washed with PBS and then replaced with HBSS (containing calcium, magnesium ions and 10mM Hepas buffer) medium for 2, 3, 4 hours respectively , as a model of cell starvation. After the induction, the cells in each group were collected, and after lysing, the changes of LC3 were determined by western blotting. The result is as Figure 6 As shown, with the increase of induction time, the expression of LC3-II protein inc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com