Network construction and analysis method for oxidized bacterium gluconicum genome scale metabolism
A technology for oxidizing glucose and genome scale, applied in data visualization, special data processing applications, instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
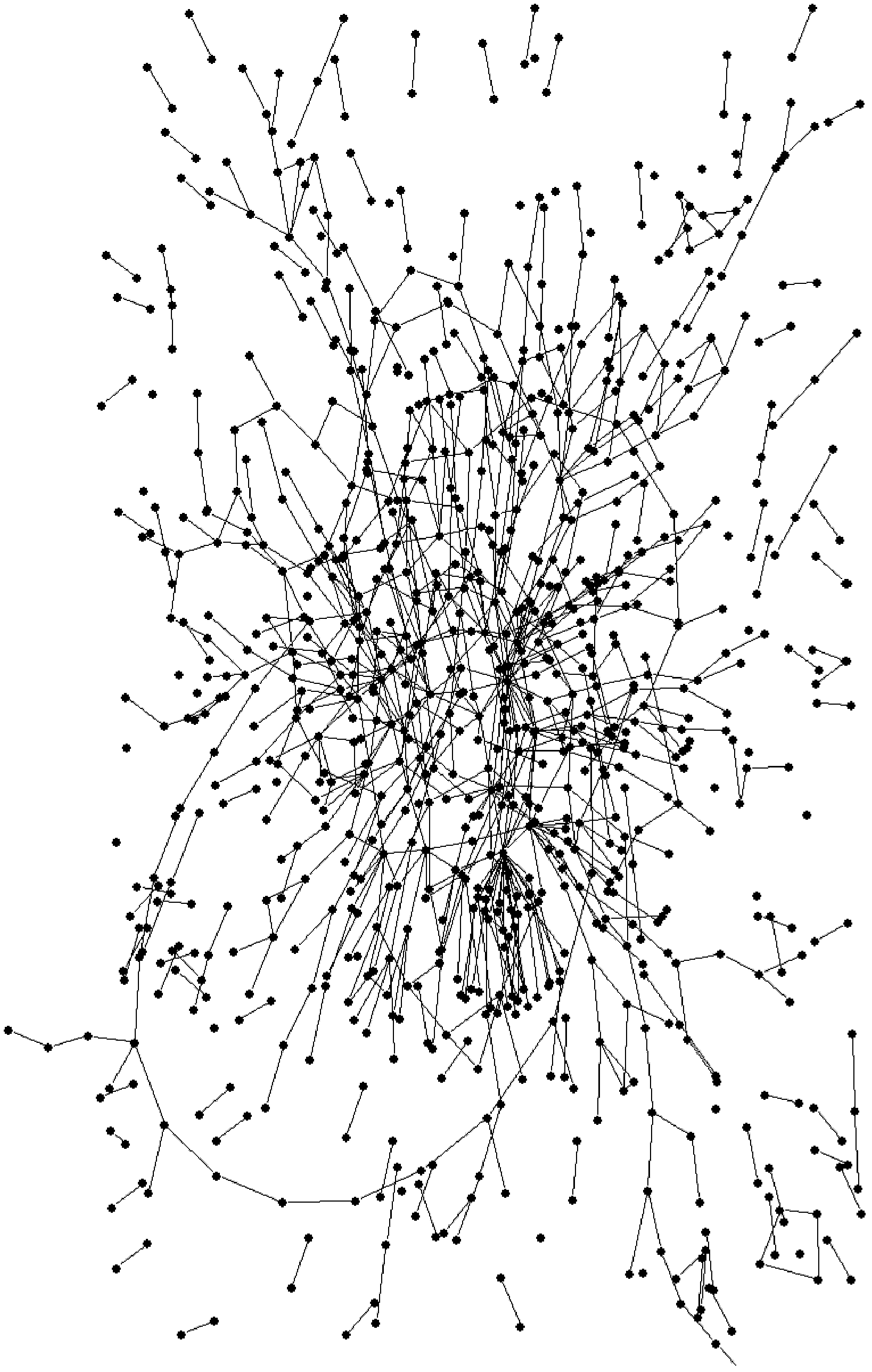
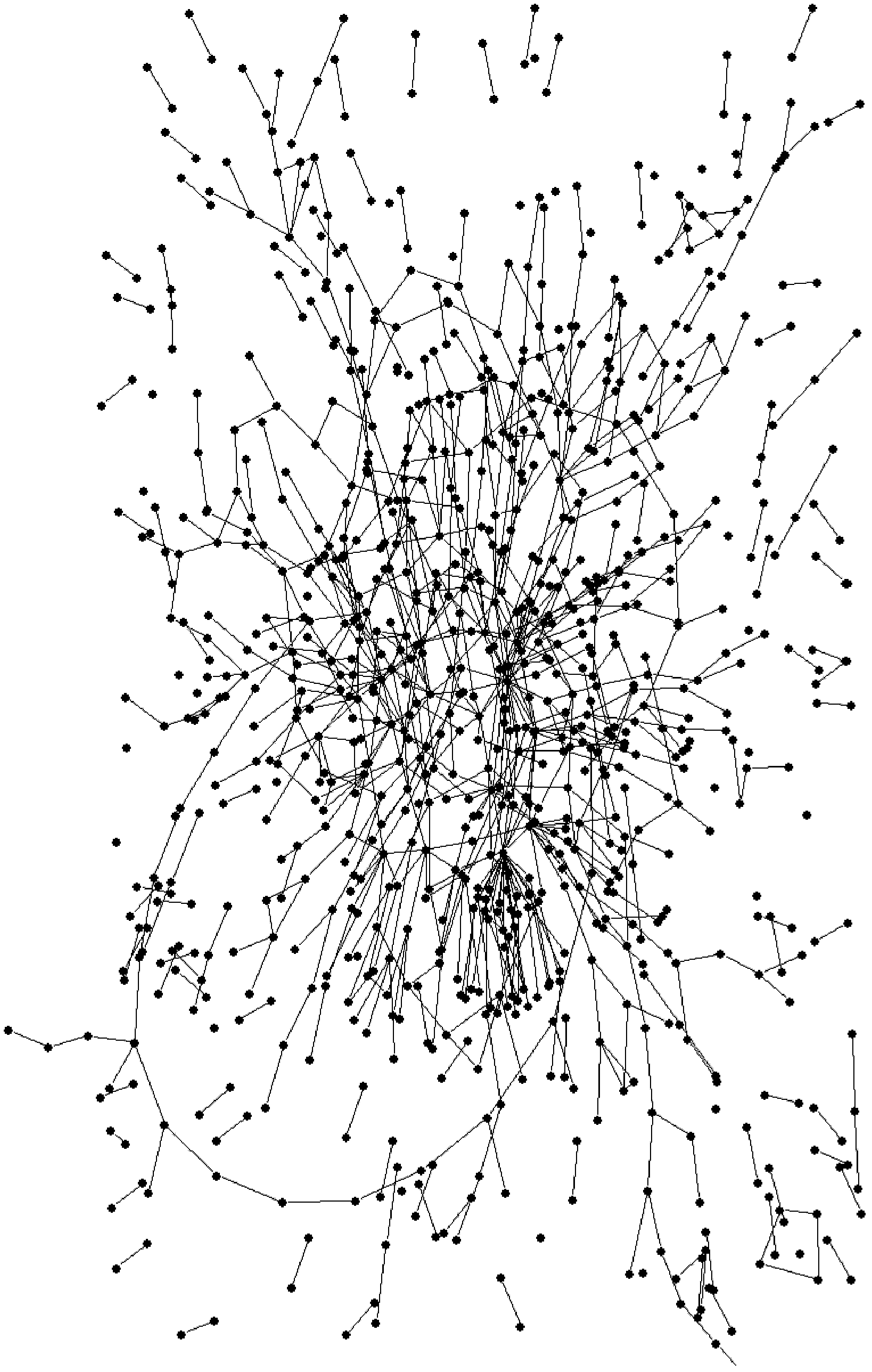
[0038] The genome-scale metabolic network of Gluconobacter oxidans was constructed according to the KEGG database and other related databases and literature reports. The metabolic network consists of 1007 metabolites, 860 reactions, 433 genes, and 403 enzymes, classified into 72 sub-metabolic pathways or subsystems, which include intracellular and extracellular small molecule metabolic reactions, biomass synthesis reactions, and transport reactions .
[0039] To use Pajek to draw a metabolic network diagram, the metabolic network needs to be modified and converted into a format recognizable by Pajek. The modification is as follows: ATP, ADP, NADH, NAD+, NADPH, NADP+, THF, and other circulating metabolites are deleted, because although they participate in a large number of metabolic reactions, they do not directly participate in the conversion process of substrates to products. Delete polymerization reactions and reactions involving macromolecules, such as synthesis of protein...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com