Primers and method for cross primer isothermal amplification detection of acidovorax citrulli
A technology for detection of melon fruit spot bacteria and constant temperature amplification, which is applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of time-consuming and expensive instrument configuration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Design and synthesis of embodiment 1 primer
[0029] Five primers were designed according to the 16S rDNA sequence of Melon fruit spot pathogen, and the primer sequences are:
[0030] ACLF3: 5'-GGCTAACTACGTGCCAGC-3'
[0031] ACLB3: 5'-ACGCATTTCACTGCTACA-3'
[0032] ACLBIP: 5'-CAGATGTGAAATCCCCGGGCTCTGCCGTACTCCAGCGAT-3'
[0033] ACDF5b1: 5'-GCAAGCGTTAATCGGAATTACT-3'
[0034] ACDF5f2: 5'-CAACCTGGGAACTGCATTTGT-3'
[0035] Among them, ACLF3 and ACLB3 are replacement primers, ACLBIP is a cross primer, ACDF5b1 is a forward detection primer with 5-terminal biotin-labeled, and ACDF5f2 is a reverse detection primer with 5-terminal FITC-labeled.
Embodiment 2
[0036] The establishment of embodiment 2 cross primer amplification method
[0037] Using the total DNA extracted from a commercial plant DNA extraction kit or the isolated bacterial liquid as a template, the 20 μl reaction system contains: 0.8 μmol / L cross primer ACLBIP; 0.1 μmol / L forward detection primer ACDF5b1, 0.3 μmol / L L reverse detection primer ACDF5f2, two displacement primers ACLF3 and ACLB3 at 0.1 μmol / L each, 0.4 mmol / L dNTP, 2 μl 10× reaction buffer; 8 units of Bst DNA polymerase, 2 mmol / L MgSO 4 , and 4 μl of target DNA.
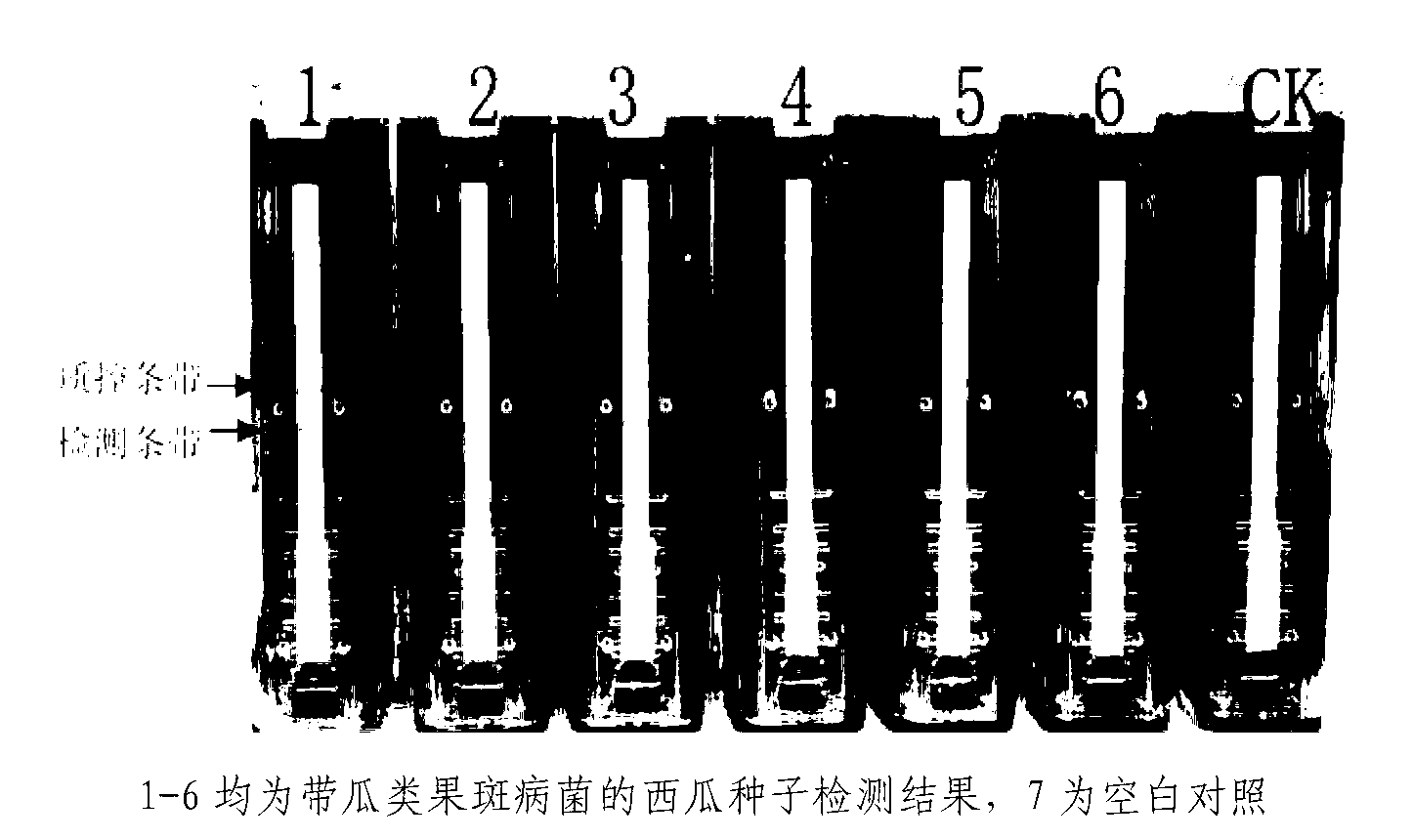
[0038] The amplification reaction was at 63° C. for 60 min, and then the amplification product was detected. The results of the amplification products are interpreted by a commercial disposable fully enclosed rapid detection device for target nucleic acid amplification products. Interpret the results by observing the color development of the detection strip and the quality control strip of the test strip. If both the detection band and the ...
Embodiment 3
[0039] Embodiment 3 cross primer amplification method sensitivity determination
[0040] The melon fruit blight bacteria suspension quantitatively counted by plate culture was diluted tenfold, and 2 μl of the bacterial pure culture of each gradient was used as a reaction template to test the detection sensitivity of the method. After testing, the tested 3.7× 10 2 The detection band and the quality control band can be observed in the bacteria solution with a concentration above cfu / ml, while 3.7×10 2 There are only quality control strips below the cfu / ml concentration, so the sensitivity of the method is determined to be 3.7×10 2 cfu / ml, because 2 μl was added to each reaction, the lowest sensitivity after conversion was 7.4 bacteria.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com