Detection method of single nucleotide polymorphism and molecular markers of cattle FBXO32 genes
A technology of single nucleotide polymorphism and molecular marker, applied in the field of molecular genetics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
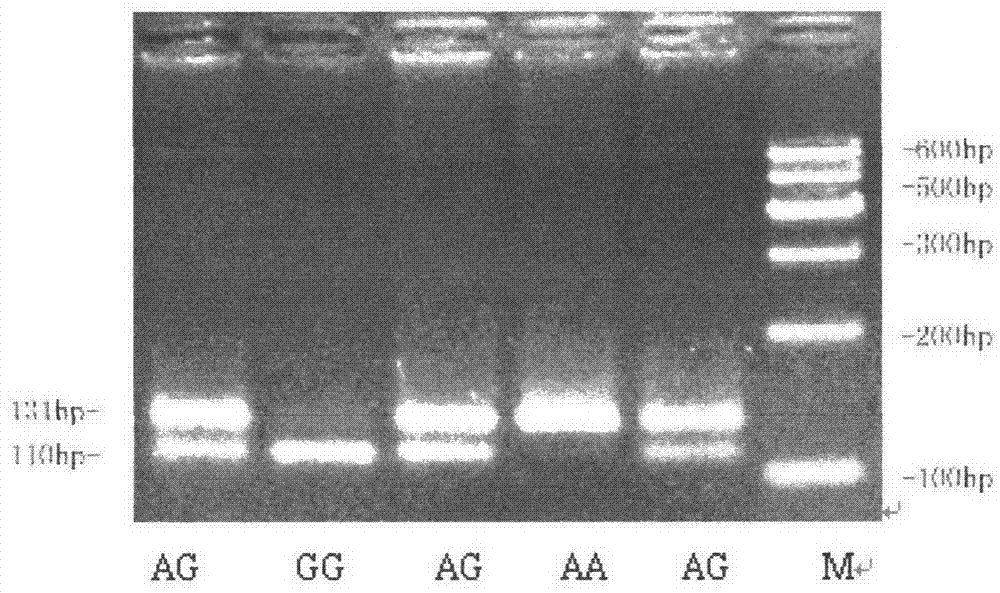
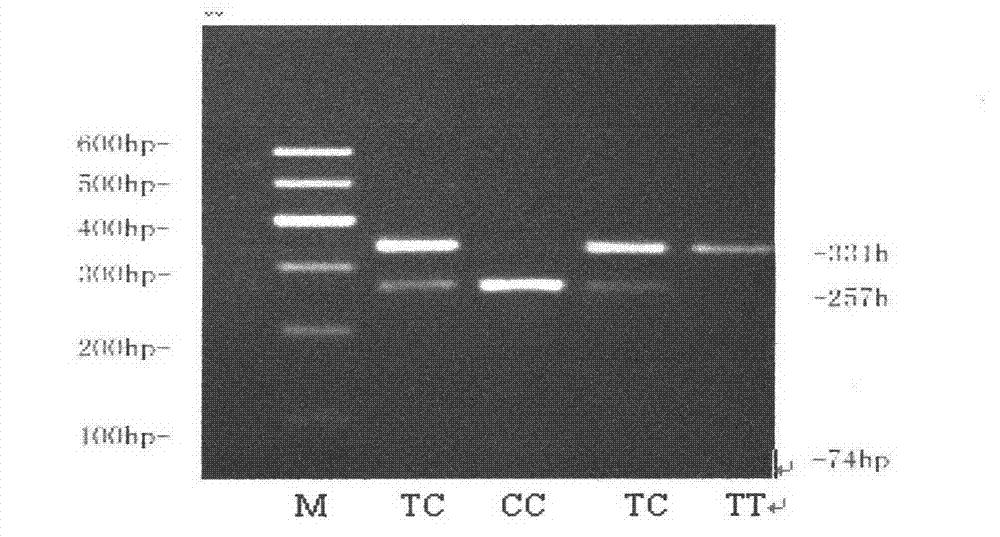
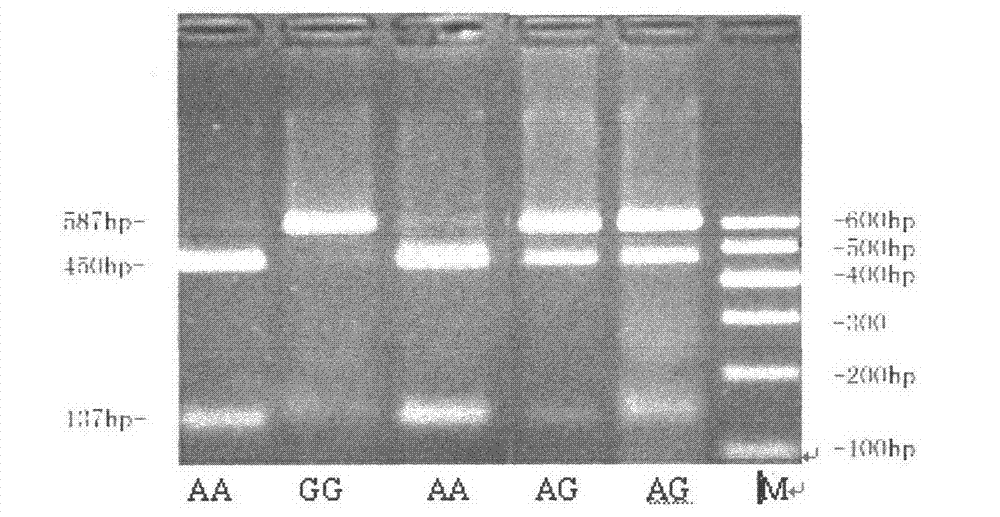
[0028] The present invention utilizes PCR-RFLP and ACRS-PCR-RFLP method to yellow cattle FBXO32
[0029] The single nucleotide polymorphisms of the 22830th, 44675th and 53423rd genes were detected
[0030] Measurement and correlation analysis, the present invention will be described in further detail below, described is to the present invention
[0031] explanation rather than limitation.
[0032] a. PCR of cattle FBXO32 gene containing exon 3, intron 8 and intron 10
[0033] Primer design
[0034] Taking the bovine sequence published by NCBI (NC_007312.4) as a reference, using Primer 5.0
[0035] The design can amplify the exon 3, intron 8 and intron 8 of the cattle FBXO32 gene respectively.
[0036] PCR primers for intron 10.
[0037] The primers for amplifying exon 3 are:
[0038] Upstream primer: 5′CGACCCTTCTACCGTGCGTTCC 3′
[0039] Downstream primer: 5′GGGCTCCCTCCTCACCCTGTT 3′
[0040] The primers for amplifying the 8th intron are:
[0041] Upstream primer: 5′GGC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com