Microarray lattice method for biological chip
A biochip and microarray technology, applied in the biological field, can solve the problems of reducing detection accuracy and achieve the effects of avoiding space effect errors, eliminating space effect errors, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
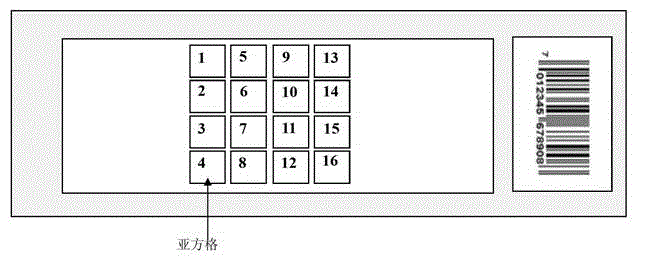
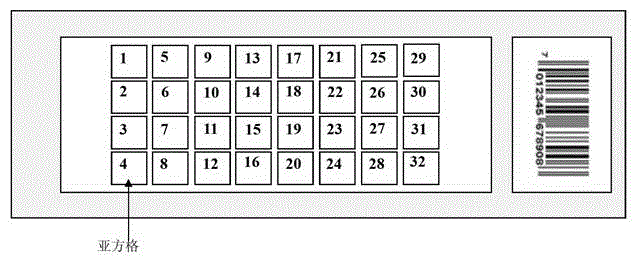
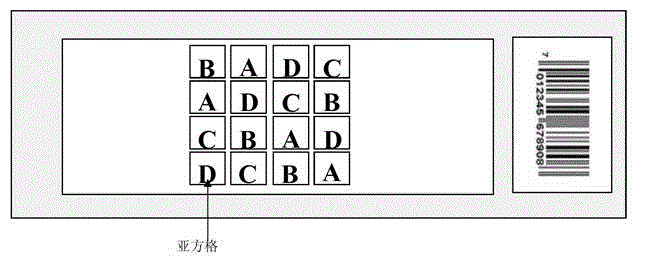
Image
Examples
Embodiment 1
[0050] Example 1. Comparison of spatial effect error absorption and digestion between the Latin square design of the antigen protein chip and the conventional design
[0051] In this embodiment, a single biological sample is used to complete the entire embodiment, which is also a unique feature of the present invention. Using the same biological sample can simplify the analysis and statistics, making the target, process and result clearer. The antigenic protein clone library was prepared according to the method of Chatterjee, a clone was taken from the clone library, and three antigenic protein biological samples of 384-well plates were prepared with this clone (these 1152 wells contained the same clone). Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, Witkin SS, Fishman D, Munkarah A, Morris R, Levin NK, Shirley NN ,Tromp G,Abrams J,Draghici S,Tainsky MA.Diagnostic markers of ovarian cancer by high-throughput antigen cloning a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com