Primer and method for detecting transgenic maize strain MIR162
A technique for transgenic corn and detection methods, applied in biochemical equipment and methods, microbe determination/inspection, DNA/RNA fragments, etc., can solve problems such as non-specific detection methods for MIR162 strains, and achieve good specificity and high sensitivity , quick and easy detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1: the detection of corn sample
[0039] 1. Primer design and synthesis:
[0040] According to the gene sequence of the transgenic maize line MIR162 (Genbank sequence number HI203349), primers were designed, consisting of forward primer 193F and reverse primer 751R, wherein the sequence of forward primer 193F is SEQ ID No: 1 and the sequence of reverse primer 751R is SEQ The specific sequence of ID No:2 is as follows:
[0041] SEQ ID No: 1: 5'-gagtcccgcaattatacat-3'
[0042] SEQ ID No: 2: 5'-ggtttgggcaaaatctcaaac-3'.
[0043] The above primers were purchased from TAKARA Company. Prepare solutions with a concentration of 10 μmol / L respectively as kits for future use.
[0044] Genomic DNA extraction:
[0045] For the DNA extraction method, please refer to "Molecular Cloning Experiment Guide, Third Edition" (written by Sambrook D.W Russell. Science Press 2002 [US] J. Huang Peitang et al. translation).
[0046] Amplification:
[0047] 3.1 PCR rea...
Embodiment 2
[0054] Example 2: Determination of the specificity of the PCR method for the detection of the transgenic maize line MIR162
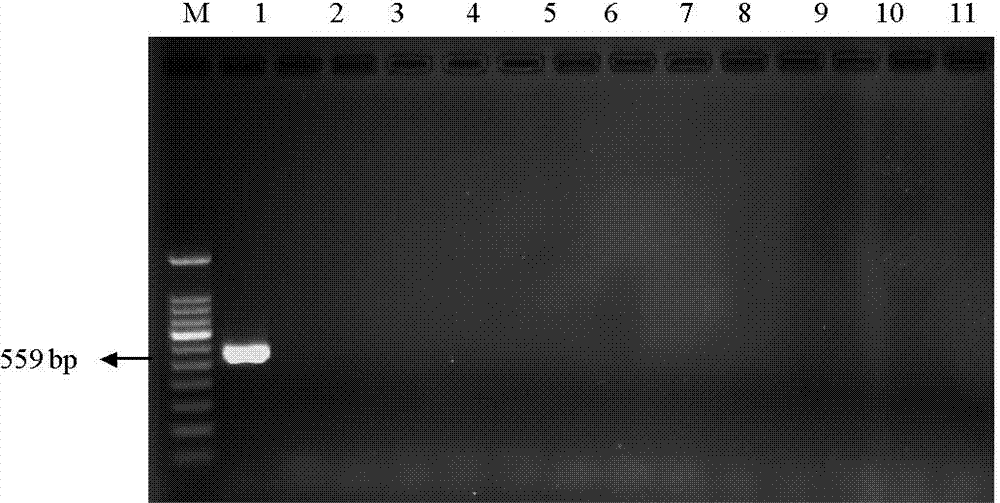
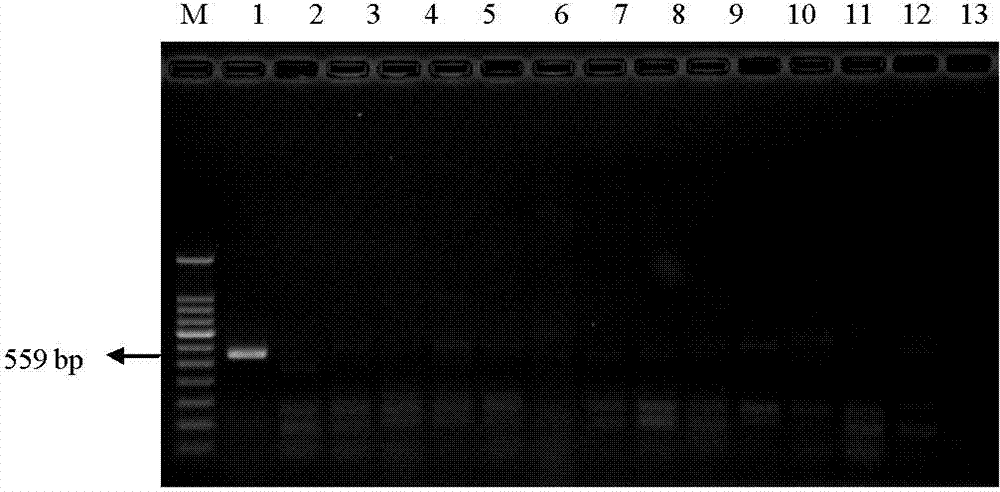
[0055] Take 13 parts of seeds or fruits of other plant species, 14 parts of standard products of different transgenic maize lines, and 1 part of non-transgenic maize. Genomic DNA was extracted according to the above method, PCR amplification was performed, and the results were detected by gel electrophoresis. The results showed that only the transgenic maize line MIR162 specifically produced an amplified band of 559 bp, and the detection results were as follows: figure 2 and image 3 shown.
[0056] See Example 1 for primers and methods used.
[0057] figure 2 It is a diagram of the experimental results of detecting MIR162 and other 13 plant species using the primers and detection method of the present invention; lanes 1-14 are sequentially transgenic corn MIR162 (10%), non-transgenic soybeans, rice, potatoes, peas, mung beans, wheat, Carrot seed...
Embodiment 3
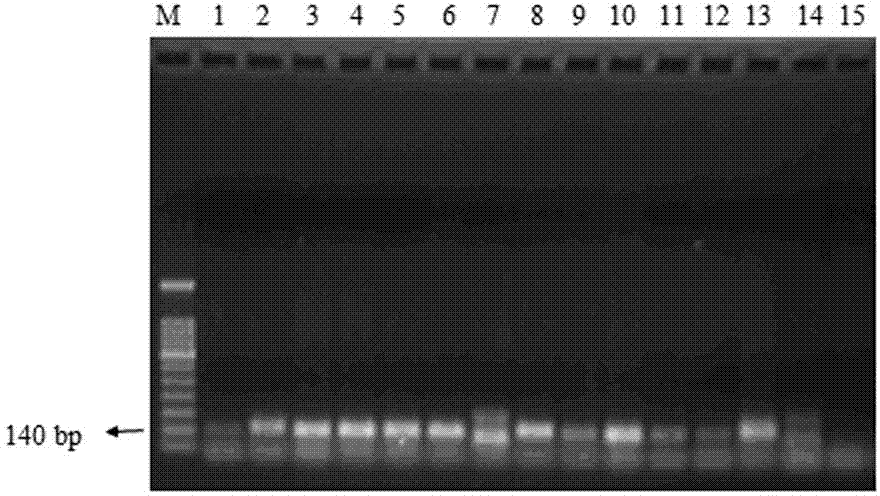
[0060] Example 3 : Sensitivity determination of a PCR method for detection of transgenic maize line MIR162
[0061] The transgenic corn line MIR162 standard was mixed with non-transgenic corn seeds to prepare the relative percentage (w / w) of the transgenic corn line MIR162 as 10%, 5%, 1%, 0.5%, 0.1%, 0.05%, 0.01% Genomic DNA was extracted respectively according to the method of the above-mentioned embodiment, and then PCR amplification was carried out, and the results were detected by gel electrophoresis. Test results such as Figure 4 shown. Figure 4 It is a diagram of the sensitivity test results of the PCR detection method of the transgenic maize line MIR162 of the present invention. Swimming lanes 1-8 are the PCR detection results of the corn seed samples and non-transgenic corn seeds with the contents of 10%, 5%, 1%, 0.5%, 0.1%, 0.05%, and 0.01% of the transgenic maize line MIR162 respectively.
[0062] See Example 1 for primers and methods used.
[0063] The exper...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com