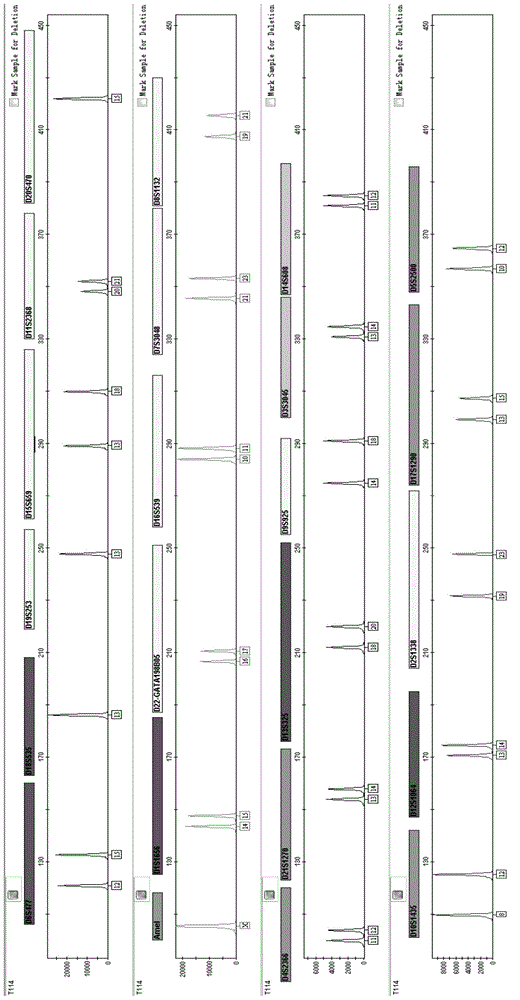
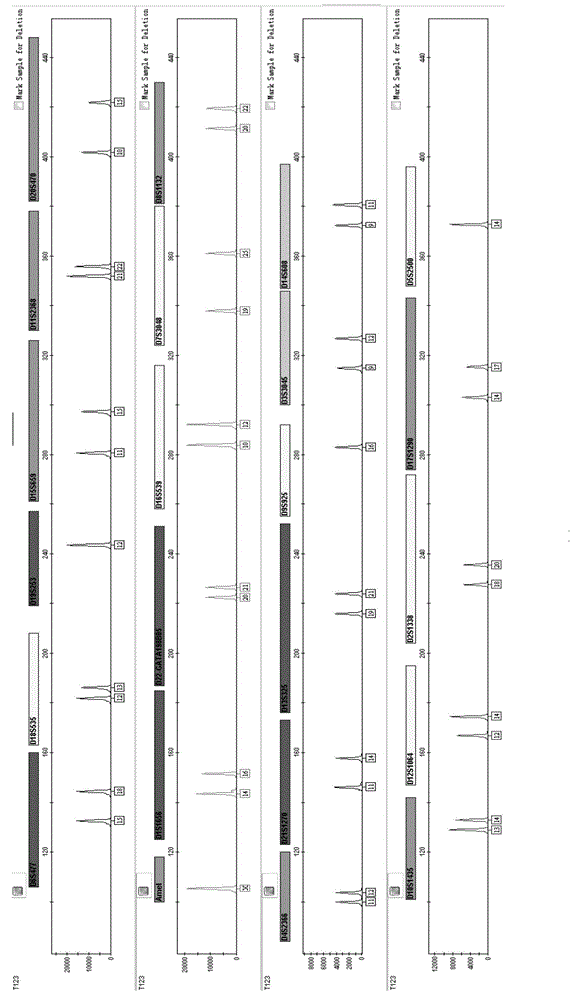
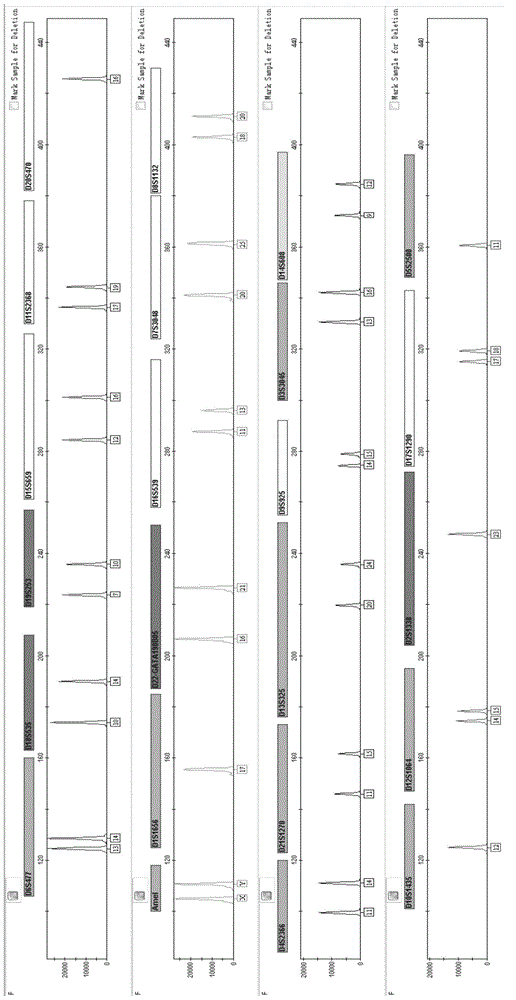
Multiplex amplification system and kit for 23 short tandem repeats
A compound amplification system and short tandem repeat technology, applied in the biological field to achieve high system efficiency and good independence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0121] Example 2 Application of 23 short tandem repeats composite amplification kit for paternity testing
[0122] 1. Collect blood samples for paternity testing (blood samples are provided by a genetic testing center)
[0123] 2. Use Chelex-100 method to extract genomic DNA (refer to "Forensic DNA Protocol". Humana Press, 1998), take 0.5-5μl of anticoagulated whole blood or 1-3mm×2-5mm blood spots into a 500μl centrifuge tube, shake Mix the Chelex solution to make the Chelex fully suspended, add 195μl Chelex-100 (5%) solution and 5μl proteinase K (20mg / ml) to each tube, shake and mix well, incubate at 56°C for two hours or overnight, take out and shake for 2 minutes, boil water After heating for 10 minutes, centrifuge at 13000 rpm for 5 minutes, and carefully transfer 150 μl of supernatant to a new centrifuge tube.
[0124] 3. Perform PCR amplification according to the amplification conditions in Example 1.
[0125] 4. Perform the test on the genetic analyzer in accordance with Exam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com