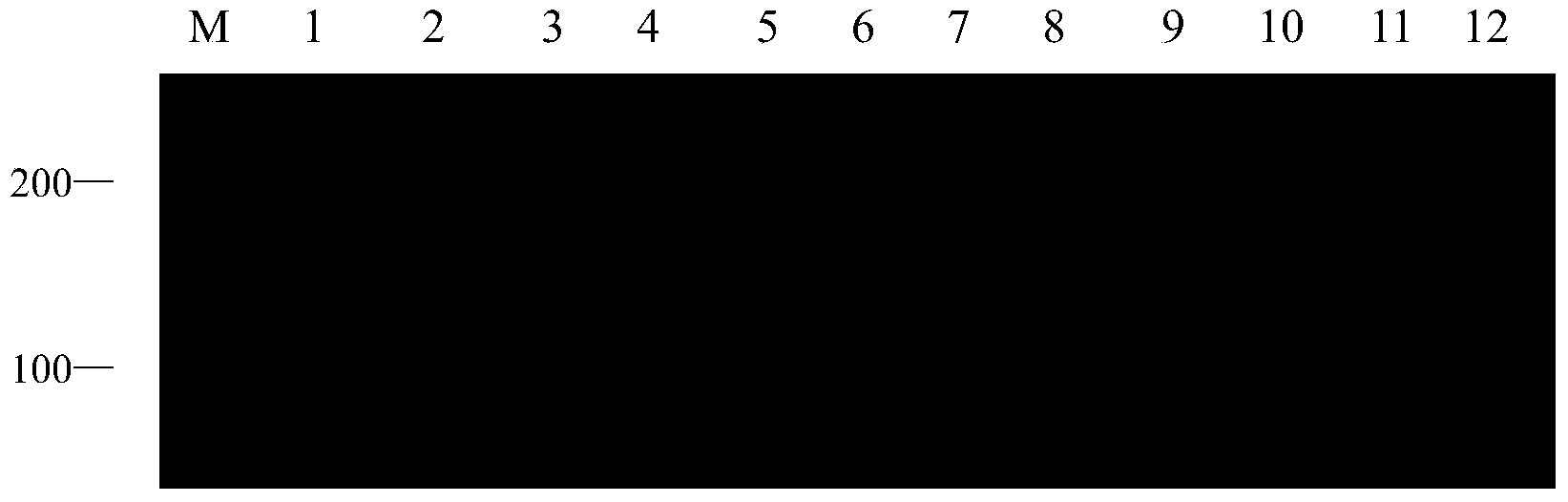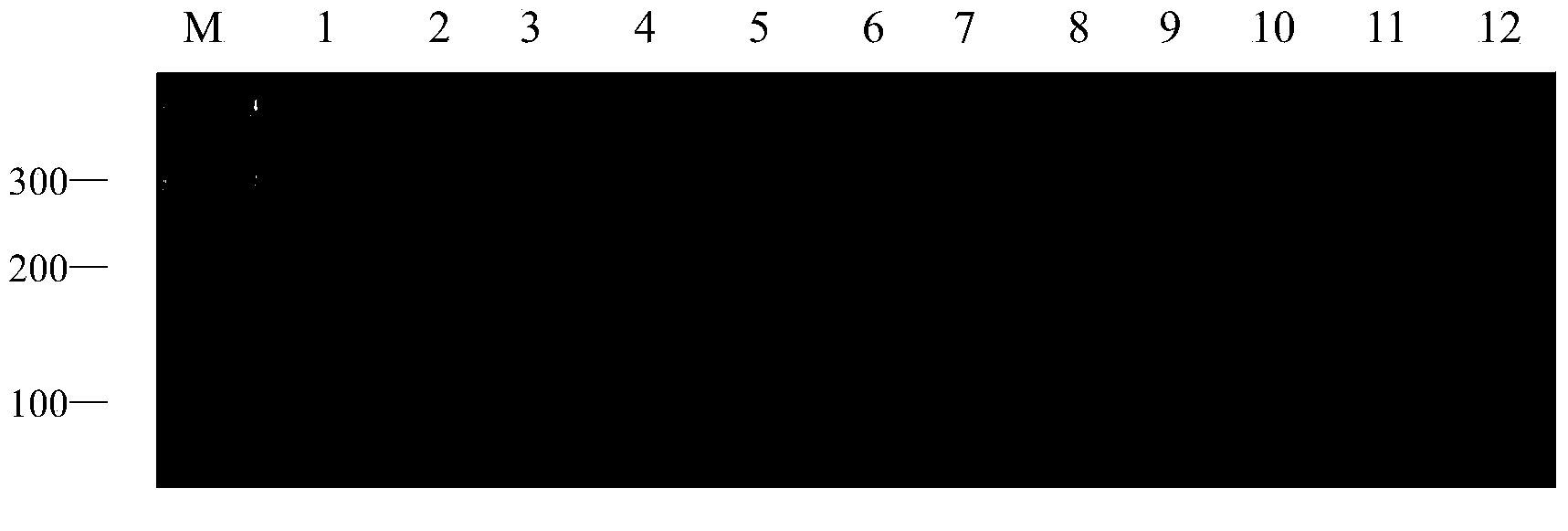Method for identifying fraxinus chinensis species by virtue of SSR (Simple Sequence Repeat) fingerprint spectrum
A fingerprint, white wax technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of protecting intellectual property rights
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
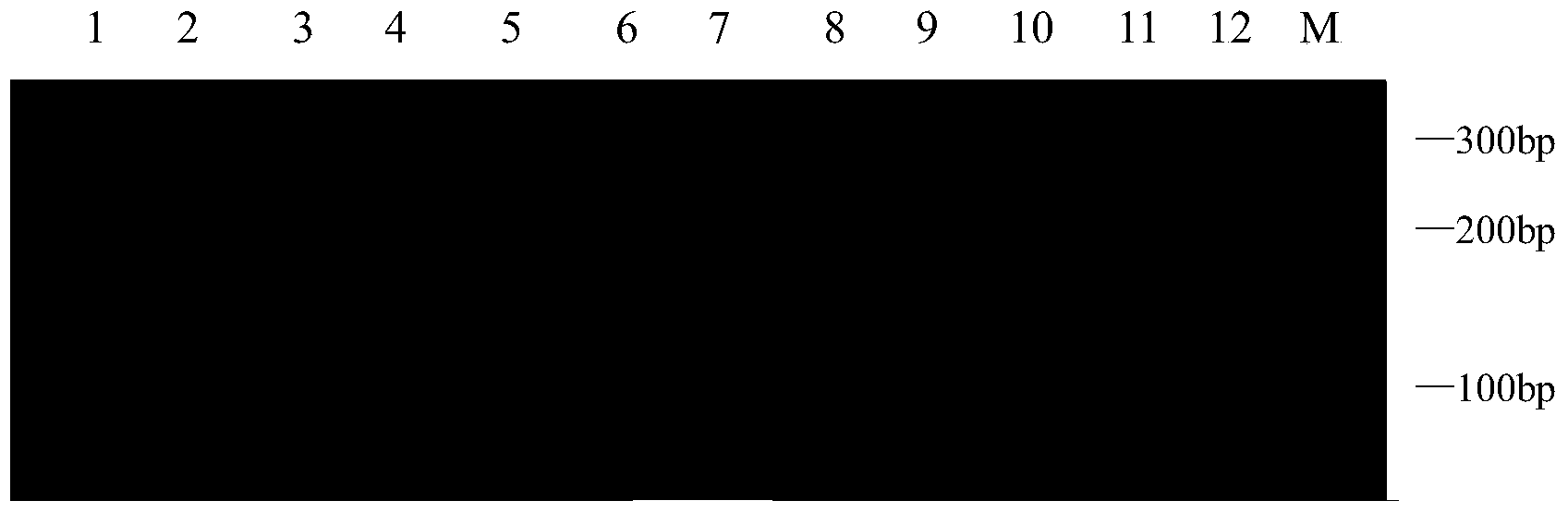
[0034] 1. Experimental materials:
[0035] In this experiment, 12 white wax core germplasms were used as test materials. They are Fraxinus sogdiana, Fraxinus chinensis Roxb, Fraxinus velutina Torr Hongyebaila, Fraxinus hupehensis, Fraxinus velutina Torr Lula No.2, Luwa Fraxinus velutina Torr Lula No.3, Fraxinus pennsylvanica Lula No.6, Fraxinus mandshurica Rupr, Fraxinus rhynchophylla, Fraxinus americana Linn , European white wax (Fraxinus Excelsior), golden leaf white wax (Fraxinus Linn). Materials were collected from Shandong Shouguang Forestry Station and Dongying Experimental Station. In May 2013, fresh leaves grown in spring were taken, packaged in fresh-keeping bags, placed in an ice box, and brought back to Shandong Forestry Research Institute. After being frozen in liquid nitrogen, they were stored in a -70°C refrigerator for later use.
[0036] 2. Genomic DNA extraction:
[0037] Take the young and healthy leaves of the test material, and use the CTAB method to ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com