Method for detecting escherichia coli in water by in-situ forming Raman enhanced substrate
A technology of Escherichia coli and Raman, which is applied in the field of in-situ formation of Raman-enhanced substrates to detect Escherichia coli in water, to achieve strong specificity, promote economic development, and ensure water safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] 1. Escherichia coli selection:
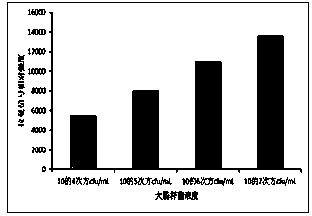
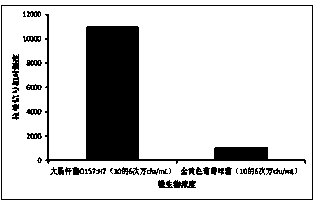
[0019] Escherichia coli O157:H7 was selected as an example to detect target bacteria. Escherichia coli O157:H7 was cultured in LB medium overnight at 37°C. Take 1mL of bacterial culture solution, centrifuge at 4000rpm / min for 5min, remove the culture medium, then use pH7.4 PBS buffer to resuspend the bacteria, dilute the bacteria, and the final concentration of the bacteria is 10 4 cfu / mL, 10 5 cfu / mL, 10 6 cfu / mL, 10 7 cfu / mL for easy detection.
[0020] 2. In situ formation of Raman enhanced substrate:
[0021] 0.1 μg / mL graphene oxide modified with 1 μg / mL glucose oxidase and 0.1 μg / mL anti-E. coli O157:H7 antibody binds to E. coli O157:H7. Centrifuge at 2000rpm / min for 5min, collect the bacteria, and resuspend in pH7.4 PBS buffer.
[0022] Add 0.5 μg / mL silver nano-seeds, glucose oxidase on the surface of graphene oxide catalyzes 1 μg / mL glucose to generate hydrogen peroxide; hydrogen peroxide reacts with 10 μg / mL silver ions ...
Embodiment 2
[0025] 1. Escherichia coli selection:
[0026] Escherichia coli O157:H7 was selected as an example to detect target bacteria. Escherichia coli O157:H7 was cultured in LB medium overnight at 37°C. Take 1mL of bacterial culture solution, centrifuge at 4000rpm / min for 5min, remove the culture medium, then use pH7.4 PBS buffer to resuspend the bacteria, dilute the bacteria, and the final concentration of the bacteria is 10 4 cfu / mL, 10 5 cfu / mL, 10 6 cfu / mL, 10 7 cfu / mL for easy detection.
[0027] 2. In situ formation of Raman enhanced substrate:
[0028] 1 μg / mL graphene oxide modified with 10 μg / mL glucose oxidase and 20 μg / mL anti-E. coli O157:H7 antibody binds to E. coli O157:H7. Centrifuge at 3000rpm / min for 5min, collect the bacterial cells, and resuspend in pH7.4 PBS buffer.
[0029] Adding 50 μg / mL silver nanoseeds, the glucose oxidase on the surface of graphene oxide catalyzes 50 μg / mL glucose to generate hydrogen peroxide; hydrogen peroxide reacts with 50 μg / mL s...
Embodiment 3
[0032] 1. Escherichia coli selection:
[0033] Escherichia coli O157:H7 was selected as an example to detect target bacteria. Escherichia coli O157:H7 was cultured in LB medium overnight at 37°C. Take 1mL of bacterial culture solution, centrifuge at 4000rpm / min for 5min, remove the culture medium, then use pH7.4 PBS buffer to resuspend the bacteria, dilute the bacteria, and the final concentration of the bacteria is 10 4 cfu / mL, 10 5 cfu / mL, 10 6 cfu / mL, 10 7 cfu / mL for easy detection.
[0034] 2. In situ formation of Raman enhanced substrate:
[0035] 0.5 μg / mL graphene oxide modified with 5 μg / mL glucose oxidase and 10 μg / mL anti-E. coli O157:H7 antibody binds to E. coli O157:H7. Centrifuge at 2500rpm / min for 5min, collect the bacterial cells, and resuspend in pH7.4 PBS buffer.
[0036]Add 30 μg / mL silver nanoseeds, and the glucose oxidase on the surface of graphene oxide catalyzes 30 μg / mL glucose to generate hydrogen peroxide; hydrogen peroxide reacts with 30 μg / mL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com