Gene joint detection method and kit
A combined detection and kit technology, applied in the field of molecular biology, can solve the problems of patient payment, a large number of clinical samples, high detection costs, etc., and achieve the effect of strong selectivity, strong specificity, and low sample consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Take 2 μL of the DNA in Table 2 and put it in a PCR tube as a template, place it on a PCR instrument, set the temperature at 95°C, maintain it for 5min, and then cool it down to 25°C. Add 1.5 μL MLPA Buffer and 1.5 μL Probemix (the Probemix contains 34 probes in Table 1) to the denatured template, and run the hybridization program on the PCR machine: 95°C for 1min; 60°C for 16h. After the hybridization reaction, add 15 μL of ultrapure water and 0.1 μL of ligase to each PCR tube, and run the ligation program on the PCR instrument: 54°C for 20 minutes; 95°C for 10 minutes; cool to 20°C. PCR amplification was performed after the ligation reaction was completed. Add 1.0 μL of PCR universal primers, 1.0 μL of dNTPs, and 0.5 μL of Taq enzyme to each PCR tube, and run the PCR program on the PCR machine:
[0045]
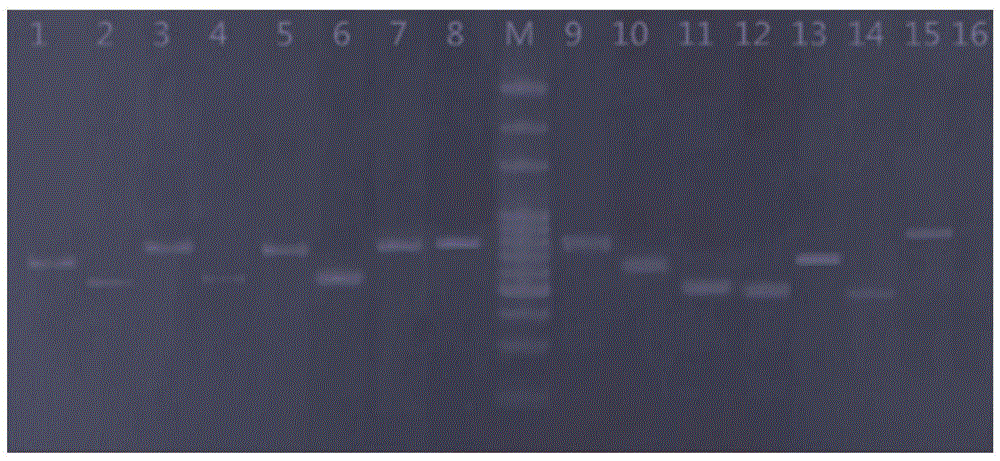
[0046] Detection of amplified nucleic acid fragments by polyacrylamide electrophoresis, see figure 1 . Each mutation point is accurately detected, and the leng...
Embodiment 2
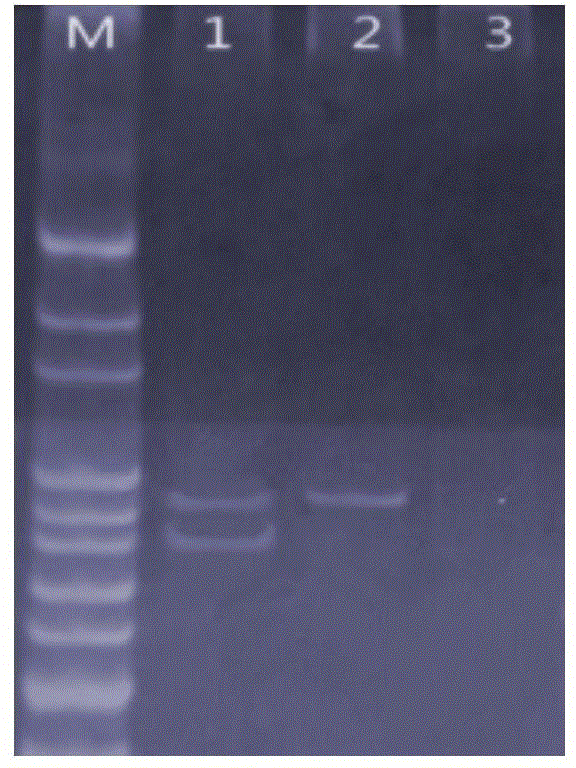
[0050] Take 2 μL of the DNA in Table 3 as a template in a PCR tube, and the rest of the inspection operations are the same as in Example 1, and the amplified nucleic acid fragments are detected by polyacrylamide electrophoresis, see figure 2 . When the DNA extracted from the blood of healthy people was used as the sample, two bands with a length of 186bp and 194bp were amplified by PCR; however, when the genomic DNA of HCC827 cells with E746-A750 deletion was used as a sample, only a band with a length of 194bp was amplified. a strip.
[0051] Table 3: Template DNA of Example 2
[0052]
Embodiment 3
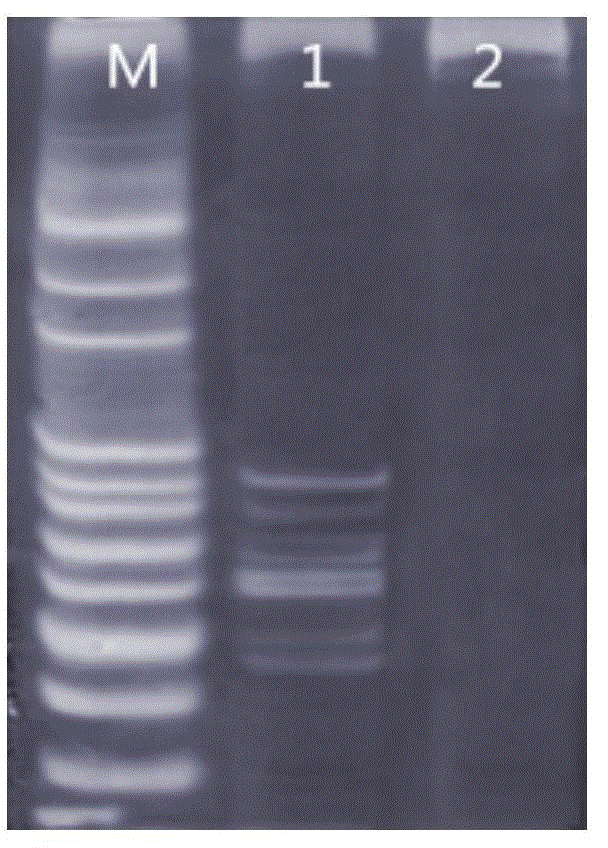
[0054] Take 2 μL of the DNA in Table 4 as a template in a PCR tube, and the rest of the inspection operations are the same as in Example 1, and the amplified nucleic acid fragments are detected by polyacrylamide electrophoresis, see image 3 . When multiple gene mutations exist in the sample at the same time, the test method can detect the mutated genes at the same time.
[0055] Table 4: Template DNA of Example 3
[0056]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com