Linked molecular marker for dominant resistance gene ZYMV-2 of cucurbita pepo L. ZYMV and application of linked molecular marker
A technology of ZYMV-2 and molecular markers, which is applied in the application field of detection of disease-resistant plants, can solve the problems of lack of zucchini ZYMV disease-resistant genes, etc., and achieve the effects of convenient identification, stable amplification, and accelerated utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
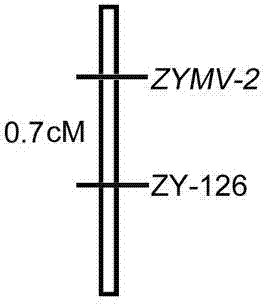
[0035] ZYMV dominant disease resistance gene in zucchini ZYMV-2 Tightly linked SSR molecular marker, said molecular marker is ZY-126, which has the DNA sequence described in SEQ ID NO. 1 and SEQ ID NO. 2 in the sequence listing.
[0036] The ZYMV dominant disease resistance gene of zucchini ZYMV-2 The method for obtaining closely linked SSR molecular markers is characterized in that it comprises the following steps:
[0037] The SSR molecular markers were obtained by using the SSR-BSA technique using the following method: using zucchini inbred lines BS11 and BS6 as susceptible and resistant parents to prepare F 2 Generation segregation population, using 584 pairs of SSR primers to screen the ZYMV dominant resistance gene in zucchini ZYMV-2 Linked molecular markers, a co-dominant molecular marker, ZY126, and disease resistance gene ZYMV-2 The genetic linkage distance of 0.7 cM.
[0038] The application of the molecular marker in the identification and transfer of ZYMV d...
Embodiment 2
[0048] This embodiment specifically describes the ZYMV dominant disease resistance gene of zucchini and zucchini ZYMV-2 A method for obtaining closely linked SSR molecular markers, comprising the following steps:
[0049] (1) Zucchini inbred lines BS11 and BS6 F 2 Generation generation and phenotyping:
[0050] (1) Hybrid F obtained by crossing zucchini inbred line BS11 (female parent) and BS6 (male parent) 1 , F 1 F 2 generation groups.
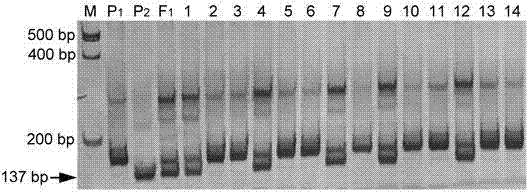
[0051] (2)F 2 The single plant of the generation group was planted in a nutrient pot in a greenhouse, covered with an insect-proof net, and inoculated with viral diseases after the cotyledons were fully unfolded, and the disease resistance traits were investigated 15 days after the inoculation. The identification results are shown in Table 1.
[0052] Table 1 BS11×BS6 F after inoculation with ZYMV virus disease 2 Generational population genetic analysis
[0053]
[0054] R and S represent resistant and susceptible plants, respec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com