Zucchini ZYMV virus disease molecular marker and method for identifying zucchini ZYMV virus disease resistance
A molecular marker, zucchini technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems of few disease-resistant varieties, long cycle, and mid-way selection of hybrid virus breeding.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1, the identification of zucchini ZYMV virus disease resistance character
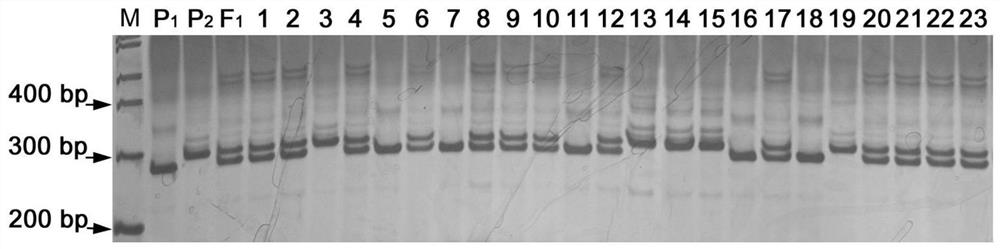
[0086] In this example, primers used to identify or assist in the identification of ZYMV virus disease resistance traits of zucchini are used to identify the resistance traits of ZYMV virus disease of zucchini to A1. A1 is composed of single-stranded DNA named P1 and P2, and P1 is SEQ ID No. The single-stranded DNA represented by positions 1-21 of 1 (P1: 7547962F, TGCACTCCAGAAGATCGTCAT), and P2 is the single-stranded DNA reverse complementary to positions 297-316 of SEQ ID No. 1 (P2: 7547962R, CTGCTTGAGCTTGCTATCCC).
[0087] 1. The zucchini to be identified are as follows, and 50 strains of each zucchini are selected for testing:
[0088] Zucchini inbred line Teizer58, whose phenotype is susceptible to ZYMV virus disease;
[0089] Zucchini inbred line Cucurbita 8 father, whose phenotype is resistant to ZYMV virus disease;
[0090] Ailu No. 1, whose phenotype is susceptible to ZYMV v...
Embodiment 2
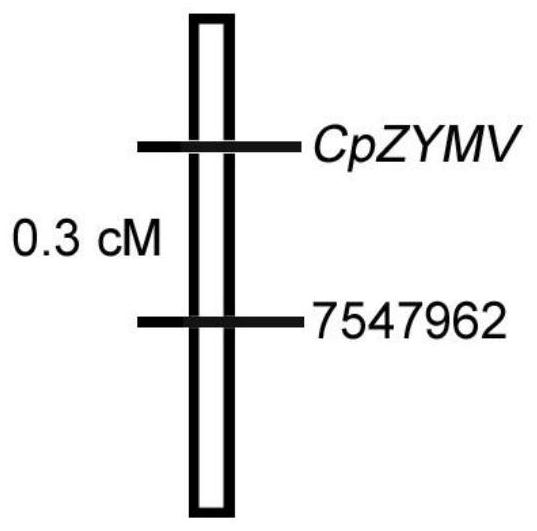
[0115] Example 2, the acquisition method of the InDel molecular marker closely linked with the zucchini ZYMV virus disease dominant resistance gene CpZYMV
[0116] The zucchini ZYMV virus disease molecular marker of Example 1 is named molecular marker 7547962, and this embodiment specifically describes the method for obtaining the InDel molecular marker 7547962 closely linked with the zucchini ZYMV virus disease dominant resistance gene CpZYMV, including the following step:
[0117] (1) Zucchini inbred line Cucurbita 8 father and Teizer58F 2 Generation generation and phenotyping:
[0118] (1) Cucurbit inbred line Cucurbita 8 father (female parent, P1) was crossed with Teizer58 (male parent, P2) to obtain hybrid F 1 , F 1 F 2 generation groups.
[0119] (2) 192 strains of F 2The single plant of the generation group was planted in a greenhouse nutrient pot, covered with an insect-proof net, and inoculated with ZYMV virus disease after the cotyledons were fully expanded, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com