Primer group complexly amplified by fluorescent mark for synchronously analyzing 27 genetic locus of human genomic DNA, kit and application
A technique of fluorescent labeling and multiple amplification, applied in the field of molecular biology, to achieve good allele frequency distribution, high genetic polymorphism, and increase the possibility of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
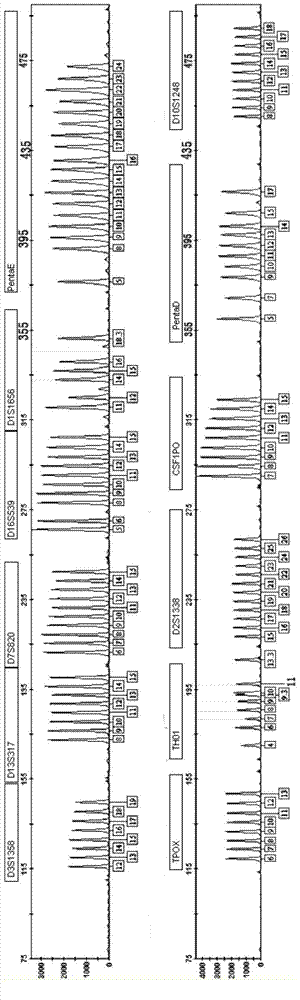
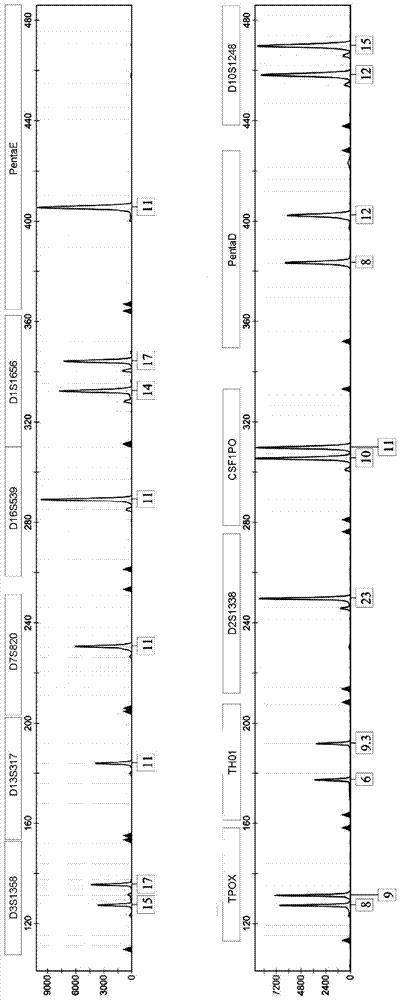
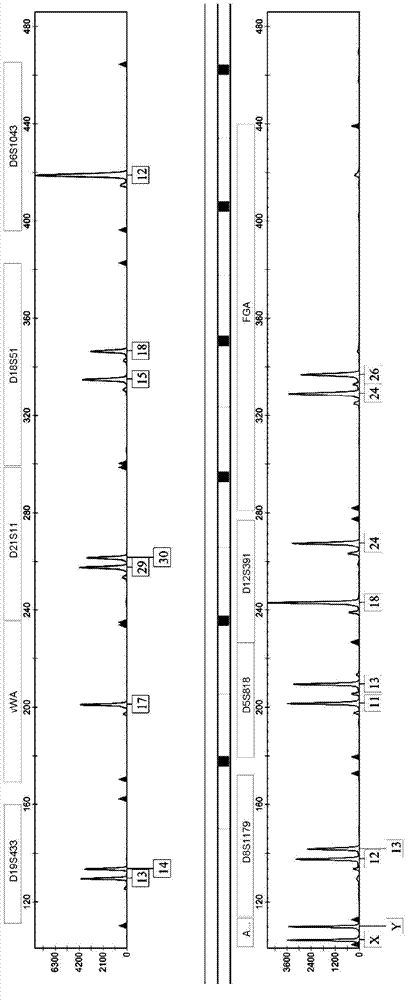
[0042] 1. Determination of genetic loci
[0043] Perform in-depth analysis of existing STR loci and optimize new high-discrimination loci. 对D1S1656、D6S1017、D4S2408、D8S1179、D21S11、D7S820、D13S317、D16S539、TPOX、TH01、D5S818、vWA、D18S51、FGA、D2S1338、D19S433、Penta E、D10S1248、D1S1677、Penta D、CSF1PO、D3S1358、D19S253、D12S391 、D6S1043、D6S474、D12ATA63、D22S1045、D11S4463、D1S1627、D3S4529、D2S441、D17S1301、D1GATA113、D18S853、D20S482、D14S1434、D5S2500、D2S1776、D10S1435和D9S1122等40多个STR基因座在中国人群的遗传多态性开展 Research. Genotype detection was performed on more than 5,000 individuals, and data such as individual recognition ability (DP), heterozygosity (H), and non-parent exclusion rate (PE) were calculated according to the distribution frequency of alleles at each locus, indicating that the 24 STR genes Among the loci, except for the TH01 and TPOX loci, the DP values of the other loci are close to 0.9, the H values are greater than 0.7, and the PE values are mostly above 0.5, which shows that they have go...
Embodiment 2
[0071] Example 2 Application of 27 loci fluorescent marker multiple amplification test system for triplet paternity identification
[0072]1. Collect blood samples in paternity testing cases: the samples in this experiment are provided by XX paternity testing agency. DNA extraction Use the Chelex-100 method to extract genomic DNA from 3 whole blood samples: take 0.5-5 μl of whole blood and place it in a sterilized 1.5ml centrifuge tube, add 1ml sdH2O to the tube, shake for a few seconds; place at room temperature for 10 minutes Afterwards, shake for a few seconds, centrifuge at 12,000rpm for 3 minutes, discard the supernatant, keep enough supernatant to cover the precipitate, do not stir the precipitate; add 200μl 5% Chelex-100, shake for a few seconds; incubate at 56°C for 30 minutes, shake A few seconds; boiling water bath for 10 minutes, shake for a few seconds; centrifuge at 12,000rpm for 5 minutes, the supernatant is the extracted genomic DNA. Genomic DNA was extracted w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com