Small RNA (ribonucleic acid) detection kit and quantitative method based on unbiased recognition and isothermal amplification
A detection kit and constant temperature amplification technology, which is applied in the direction of microbial measurement/testing, biochemical equipment and methods, etc., can solve the problems of affecting enzyme recognition ability and inaccurate detection results, and achieve rapid enzyme synergistic cascade constant temperature amplification. Increased response, increased sensitivity, and simple design effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
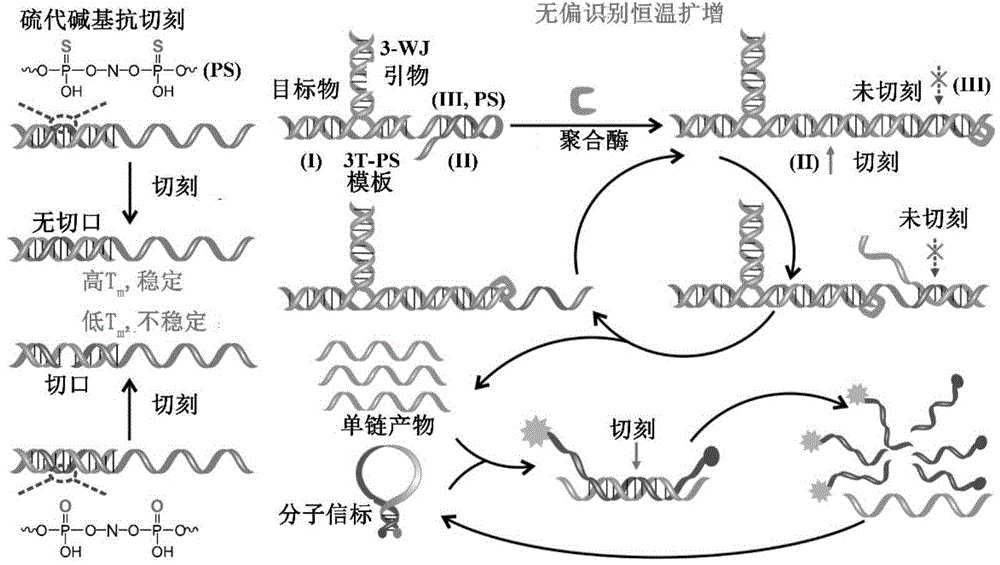
[0047] A small RNA detection kit based on unbiased recognition and constant temperature amplification, including:
[0048] A composition that forms a three-way hybrid structure with the target small RNA, including 10nM 3-WJ primer and 10nM 3-WJ template:
[0049] The 3-WJ primer consists of two parts, the 5' end part is complementary to the 3' end part of the target small RNA, and the 3' end part is complementary to the middle segment of the 3-WJ template;
[0050] The 3-WJ template is composed of three parts, the 3' end part is complementary to the 5' end part of the target small RNA, the middle part is complementary to the 3' end part of the 3-WJ primer, and the 5' end part contains two nuclease recognition sites The SDA template region of the point; one of the enzyme cleavage sites has been thio-modified;
[0051] The nucleotide sequence of the 3-WJ primer is (5'to 3'):
[0052] GTG CTC ACT CAT CCA AAA (SEQ.ID.NO.1)
[0053] Nucleotide sequence (5'to 3') of the thio-modi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com