Assay for the parallel detection of biological material based on PCR
A forward primer and reverse primer technology, applied in the field of proteins, peptides, lipids or carbohydrates in the detection sample, which can solve the problems of low sensitivity and lack of reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0104] The feasibility of the invention is demonstrated herein using two exemplary probes / P-O and positive antibodies specific for each. The target proteins in this example are epitopes specific for influenza virus and dengue virus.
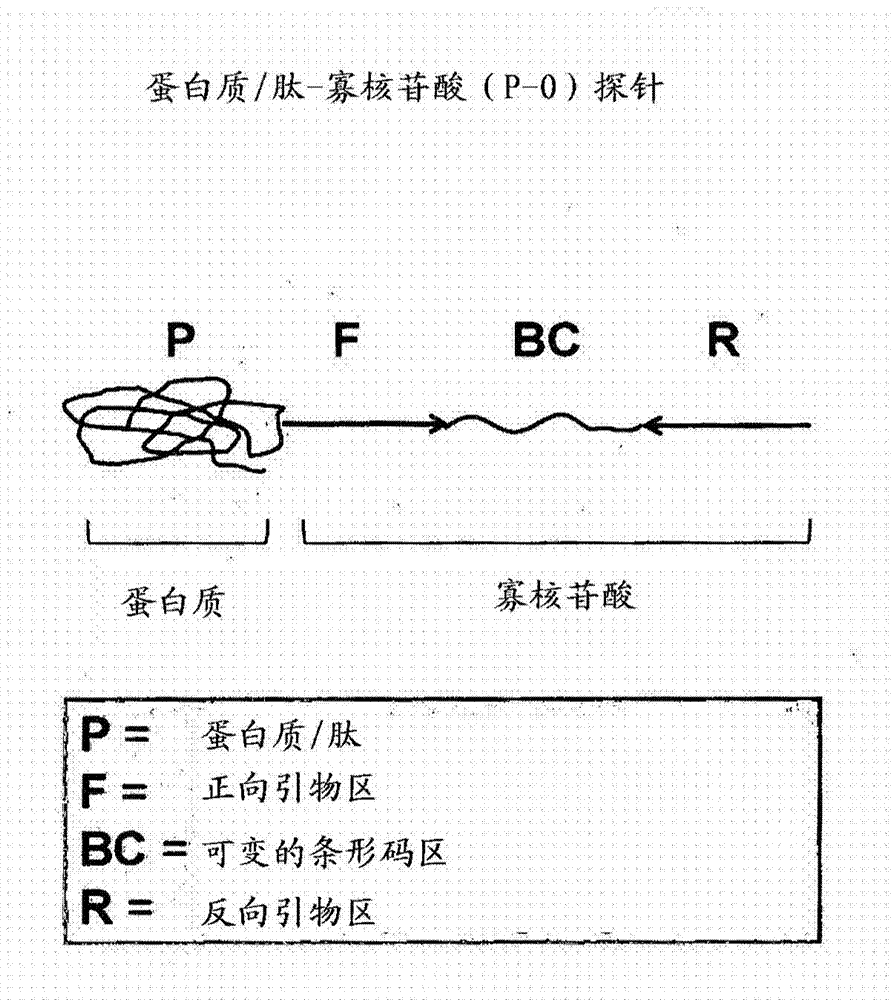
[0105] P-O conjugate and primer design (see Figure 5 ):
[0106] 1) Select two specific peptide epitopes (YPYDVPDYA and YKQPLWPNQISW, in Figure 5 Shown on the left hand side of part A): one from influenza virus and the other from dengue virus.
[0107] 2) In this particular example, specific restriction sites are incorporated into each of the different barcode regions for oligonucleotide design, i.e., different restriction sites are incorporated into each barcode region so that Use enzymatic digestion to confirm sequencing results. BamH1 was incorporated into the barcode region of the influenza-specific probe, whereas HindIII was incorporated into the barcode region of the dengue-specific probe.
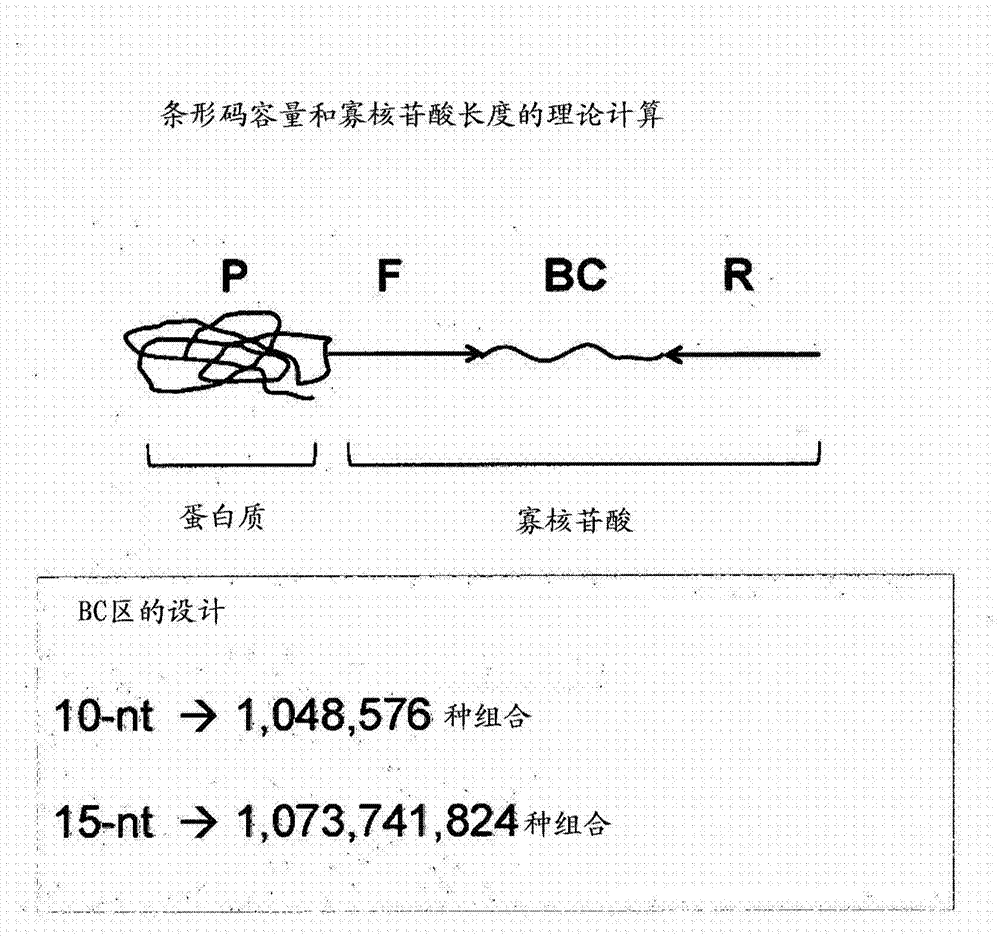
[0108] 3) In this particular example, an 8...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com