Application of nucleic acid aptamers in identifying and being bound with integrin alpha4
A nucleic acid aptamer and integrin technology, applied in the fields of biotechnology and clinical medicine, to achieve the effect of strong specificity and high affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
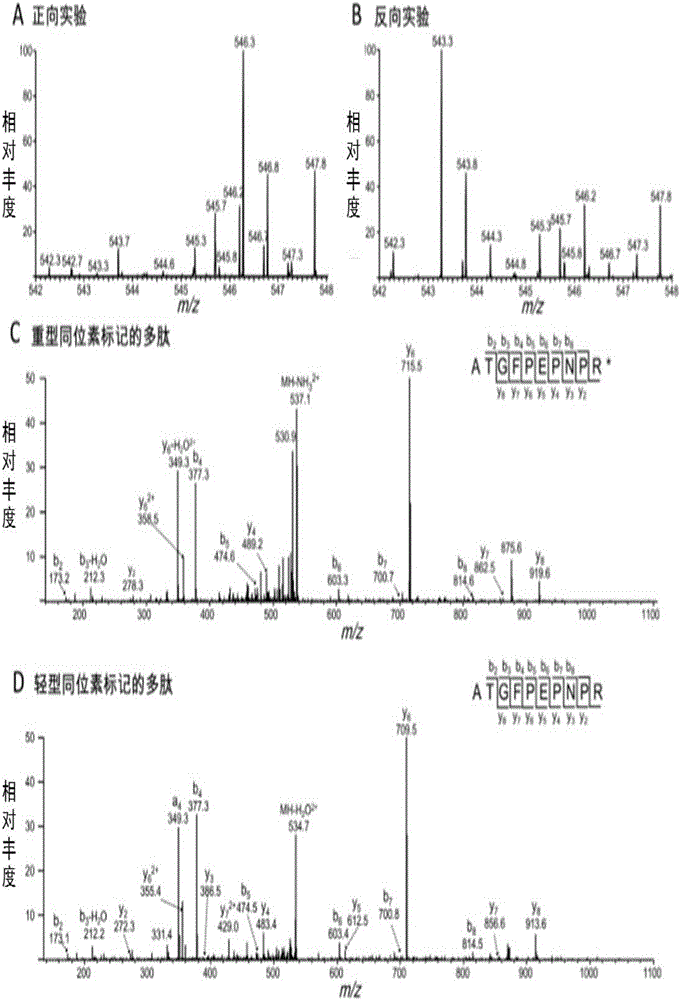
[0064] Example 1. Identification of nucleic acid aptamer Sgc-4e that specifically recognizes and binds to integrin α4
[0065] 1. Preparation of nucleic acid aptamer Sgc-4e and its derivatives
[0066] 1. Synthesis of nucleic acid aptamer Sgc-4e
[0067] Synthesize nucleic acid adapter Sgc-4e by DNA synthesizer, the nucleotide sequence of nucleic acid adapter Sgc-4e is as follows: 5'-TCACTTATTCAATTCGAGTGCGGATGCAAACGCCAGACAGGGGGACAGGAGA TAAGTGA-3' (sequence 1); The 5' end or 3' end of Sgc-4e is labeled with different molecules (such as biotin and fluorescein, etc.) to obtain derivatives of the nucleic acid aptamer Sgc-4e.
[0068] 2. DNA deprotection
[0069] After deprotection with cold ammonia, the DNA was then dissolved in TEAA solution.
[0070] 3. DNA purification
[0071] Purify by PAGE or HPLC.
[0072] 4. DNA drying
[0073] Concentrate to dryness by centrifugation.
[0074] 5. Dissolve and measure the concentration for later use.
[0075] 2. Identification of n...
Embodiment 2
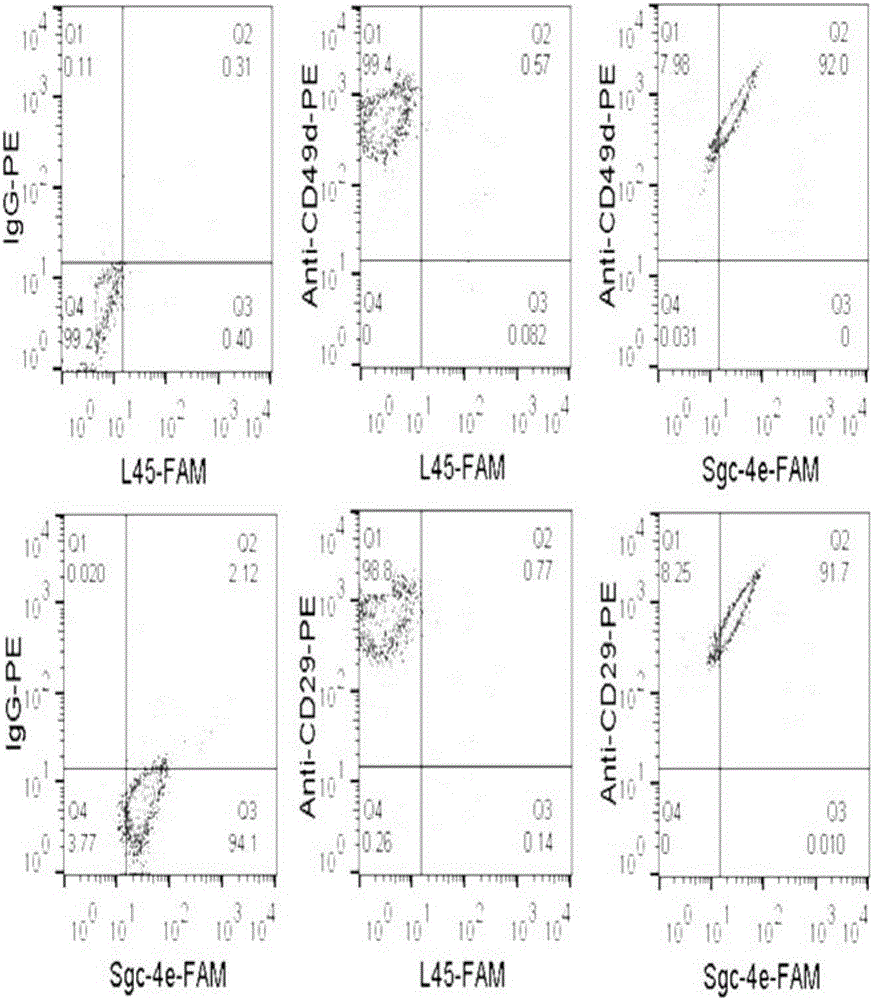
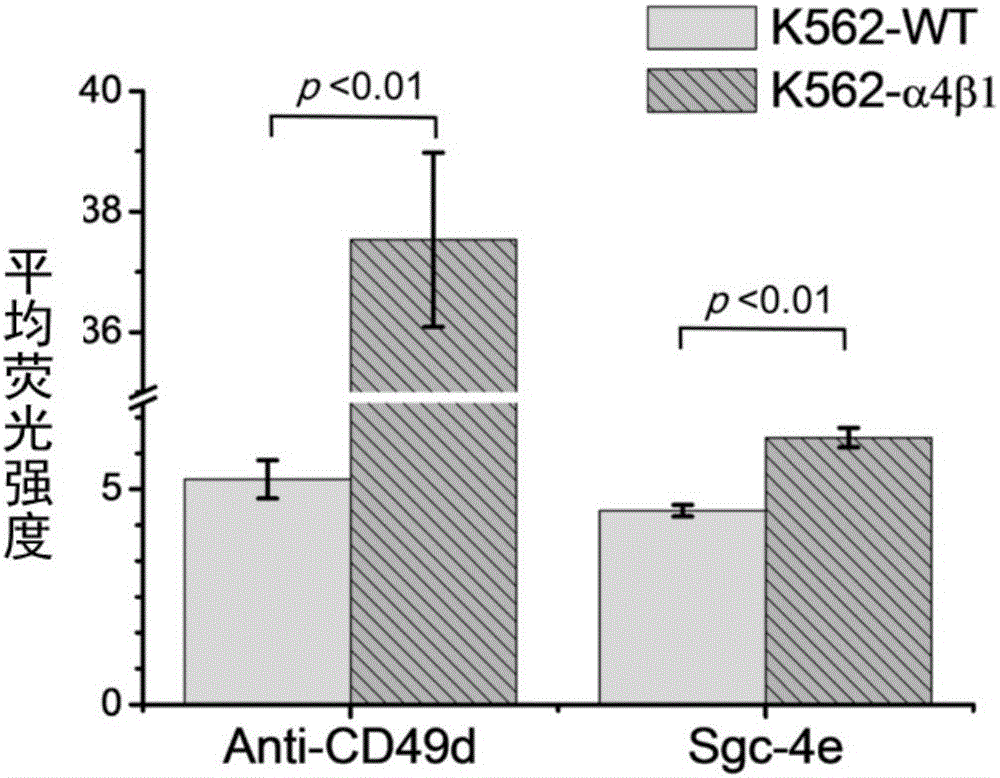
[0143] Example 2. Application of the nucleic acid aptamer Sgc-4e in determining the expression of integrin α4 in different types of cells
[0144] In order to clarify the expression of integrin α4 (NP_000876) in different cells by nucleic acid aptamer Sgc-4e, the fluorescently labeled nucleic acid aptamer Sgc-4e and anti-integrin α4 antibody were used to measure the expression of different cells by flow cytometry. The expression of integrin α4 in cells was measured. Specific steps are as follows:
[0145] 1. Preparation of fluorescein-labeled aptamer Sgc-4e solution (Sgc-4e-FAM)
[0146] The fluorescein-labeled nucleic acid aptamer Sgc-4e was obtained by coupling the fluorescein group to the 5' end of the nucleic acid aptamer Sgc-4e. Sgc-4e-FAM was dissolved in binding buffer, and the concentration was calibrated according to the ultraviolet absorption ( 100nM), heated at 95°C for 5min, placed on ice for 5min, and placed at room temperature for 15min.
[0147] 2. Pretreatme...
Embodiment 3
[0156] Example 3. Effects of divalent ions on the binding of nucleic acid aptamer Sgc-4e to integrin α4
[0157] The activity of integrin α4 is affected by divalent ions (especially Mg 2+ , Ca 2+ or Mn 2+ ) regulation, in order to prove that the nucleic acid aptamer Sgc-4e can bind to the active integrin α4, the effect of divalent ions on the binding of the nucleic acid aptamer Sgc-4e to the integrin α4 was studied.
[0158] 1. Preparation of fluorescein-labeled aptamer Sgc-4e solution (Sgc-4e-FAM)
[0159] The fluorescein-labeled nucleic acid aptamer Sgc-4e is obtained by coupling the fluorescein group to the 5' end of the nucleic acid aptamer Sgc-4e. The Sgc-4e-FAM is dissolved in the binding buffer, and the concentration is calibrated according to the ultraviolet absorption. After 100nM, heat at 95°C for 5min, place on ice for 5min, and place at room temperature for 15min.
[0160] 2. Treat the above-mentioned Jurkat E6-1 cells according to the following five groups:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com