Serum miRNA relevant to chronic heart failure and application of serum miRNA
A chronic heart failure, hsa-mir-665 technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, microbial measurement / inspection, etc., can solve the lack of detection and comparison of miRNAs in myocardial tissue, lack of epidemiological follow-up studies, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1. Screening results of miRNAs in peripheral blood and myocardial tissue of patients with chronic heart failure
[0026] 1. Collect peripheral blood samples from 10 cases of chronic heart failure (taken from inpatients in the Department of Cardiovascular Medicine from 2007 to 2014, each case was diagnosed and classified by two or more clinicians) and 10 normal controls. After sampling, centrifuge at 3,500 rpm for 6 minutes at room temperature, take the upper layer of plasma, and store it in a -80°C refrigerator. Add 1ml TRIZOL LS (Invitrogen Company) to every 0.25ml peripheral blood plasma, extract RNA, and RNasey Mini Kit (Qiagen) processes the sample. use ND-1000 detects RNA quality. miRCURY TM After being labeled with Array Power labeling kit (Exqion), the hybridization experiment was performed at the hybridization workstation. Axon GenePix 4000B microarray scanner scans to obtain chip images. GenePix pro V6.0 software analyzed the obtained data, and u...
Embodiment 2
[0029] Example 2. Analysis of miRNAs chip expression profile in peripheral blood and myocardial miRNAs chip expression profile in patients with chronic heart failure
[0030] The obtained microarray data were analyzed and processed by bioinformatics analysis methods.
[0031] 1. The scatter plot reflects the repeatability between chips or groups. The better the repeatability, the closer to the diagonal. In addition, the Pearson correlation coefficient was used to measure the repeatability, and the closer the correlation coefficient was to 1, the better the repeatability. Figure 1A with 1B The results show that the chips are finished with reliable quality and good repeatability.
[0032] 2. For comparison between groups, see Figure 1C with 1D . Use -log(Pvalue) as the ordinate and log2(Fold change) as the abscissa to plot each miRNA, and you can visually see the miRNAs with significant differences (gray dots represent miRNAs screened by Fold change and P-value) . Figur...
Embodiment 3
[0033] Example 3. Comparison of miRNAs expression profile in peripheral blood and myocardial tissue miRNAs expression profile
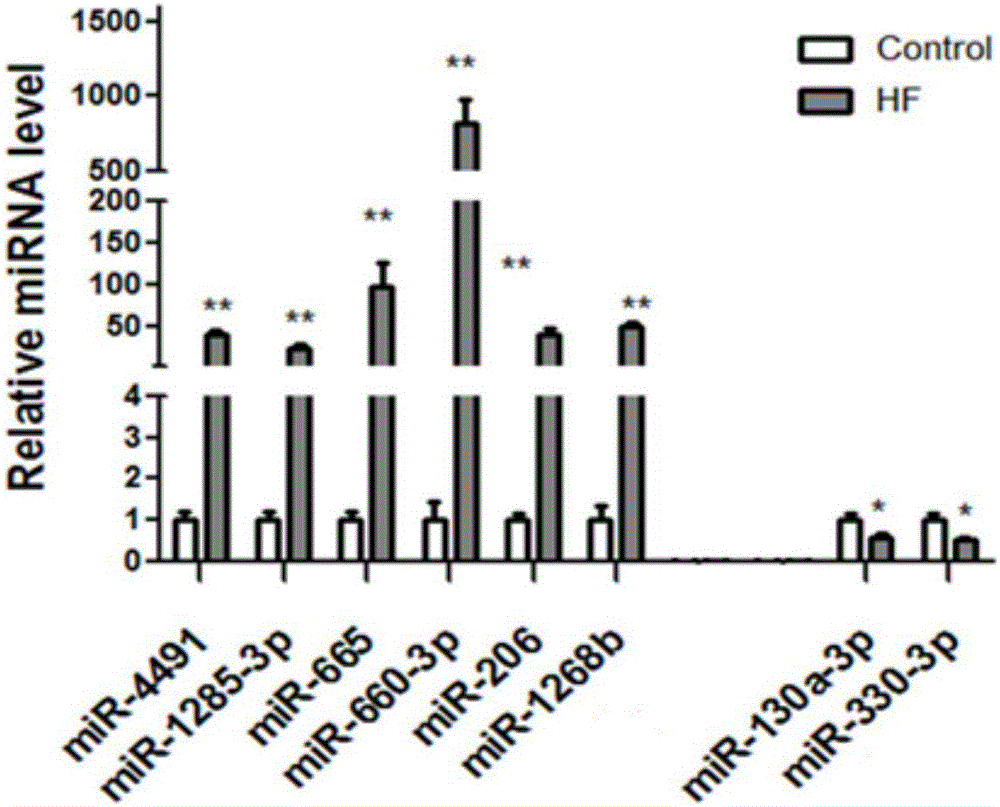
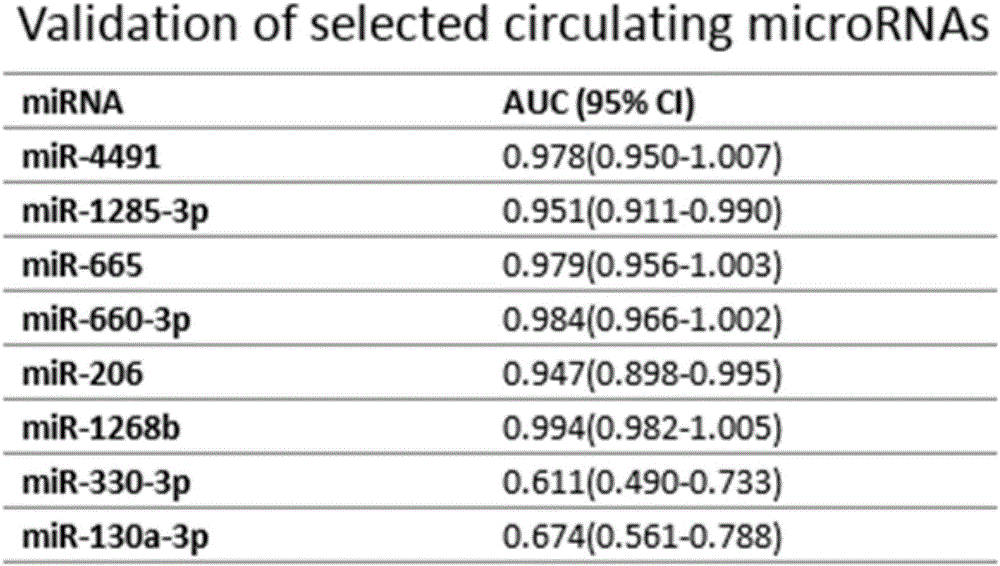
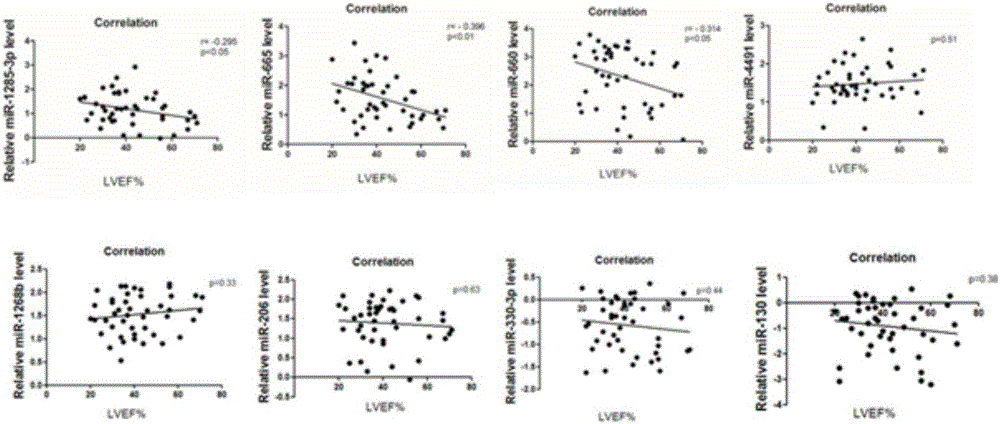
[0034] The trends of all miRNAs differentially expressed as candidate biomarkers for the diagnosis of chronic heart failure obtained from peripheral blood microarray and myocardial tissue microarray screening were classified. The results showed that there were 74 miRNAs with significant differences (including increase or decrease) in myocardial tissue expression, 599 miRNAs with significant differences (including increase or decrease) in peripheral blood, and 25 miRNAs with significant differences in expression (including increase or decrease). The expression of miRNAs in the two chips were all changed, and the expression of 6 miRNAs were all increased ( Figure 2A ) (hsa-miR-4491, hsa-miR-1285-3p, hsa-miR-665, hsa-miR-660-3p, hsa-miR-206 and hsa-miR-1268b), there were 2 species whose expression decreased ( Figure 2B ) (hsa-miR-130a-3p and hsa-miR-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com