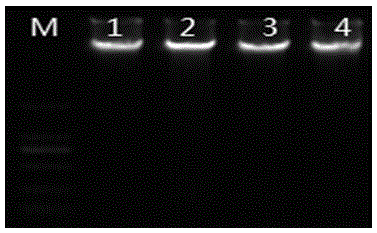
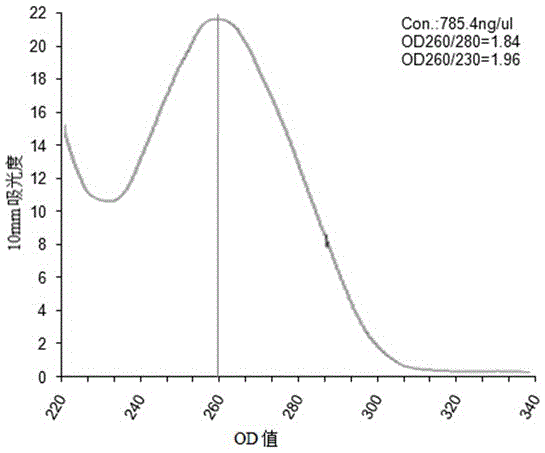
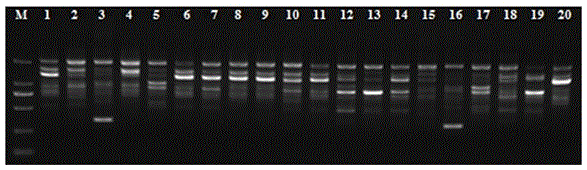
Anoectochilus roxburghii DNA extracting method suitable for RAPD analysis
An extraction method and technology for taking supernatant, applied in the field of molecular biology, can solve problems such as affecting experimental results and RAPD instability, and achieve the effect of clear bands and high repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] (1) An appropriate amount of sample (0.5g), ground with liquid nitrogen, poured into a 2mL centrifuge tube, and added CTAB solution (3%CTAB; 100mmol / L Tris-HC1, pH 8.0; 20mmol / LEDTA, pH 8.0; 2.0 mol / L NaCl; 4% PVP (K-30); before use, add 2% β-mercaptoethanol, shake, put in a 65°C water bath for 45 minutes, slightly invert several times every 5 minutes in the middle, at 20°C at 12000 rpm Centrifuge for 15 minutes.
[0027] (2) Take the supernatant, add 2 / 3 volume of chloroform: isoamyl alcohol (volume ratio 24:1), lie flat for 5 minutes, and then centrifuge at 12000 rpm for 10 minutes at 20°C.
[0028] (3) Take the supernatant, add 1 / 3 volume of chloroform, lie flat for 5 minutes, and centrifuge at 12,000 rpm for 10 minutes at 20°C.
[0029] (4) Take the supernatant, add an equal volume of -20°C pre-cooled isopropanol, add 1 / 5 of the supernatant volume 3mol / L NaAc (pH 5.2), place it in the -20°C refrigerator for 30min, Centrifuge at 12,000 rpm for 15 min to precipitate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com