Method for rapidly identifying mirror carps
A mirror carp, rapid technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as pollution, inability to identify mirror carp genome, etc., to increase feasibility and accuracy. and reliability, improve work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
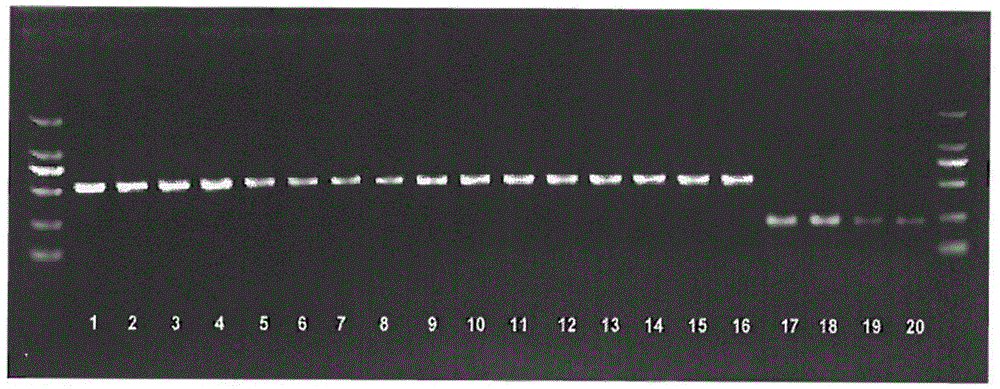
[0030] The gene sequences of mirror carp and other carp species were measured by next-generation sequencing technology. By analyzing the above-mentioned genome sequencing data and comparing the sequence differences of various carp strains, the missing sequence was found, as shown in SEQNO: 1; the primer design was directly in the deletion The upstream and downstream 200bp of the sequence are designed, and the designed primer sequences are shown in SEQNO:2 and SEQNO:3.
[0031] The above-mentioned primers provided by the present invention have high efficiency in PCR reaction and strong specificity for the amplification of the missing sequence.
Embodiment 2
[0033] 1) Take the live fish fins of mirror carp, purse red carp, colorful carp, Yellow River carp and Xingguo red carp, set 4 replicates for each fish, use NEB DNA extraction kit to extract the DNA of the fins of the above live fish, the specific operation See the instructions of the kit for steps.
[0034] 2) using the DNA of the above-mentioned mirror carp, purse red carp, colorful carp, Yellow River carp and Xingguo red carp as templates, and carrying out PCR amplification with the following primers (shown in SEQNO: 2 and SEQNO: 3),
[0035] Upstream primer 5′-TGCACGTTACTTTTAAACACTCC-3′
[0036] Downstream primer 5′-AAAAAGCAGCATGCCTTACAA-3′
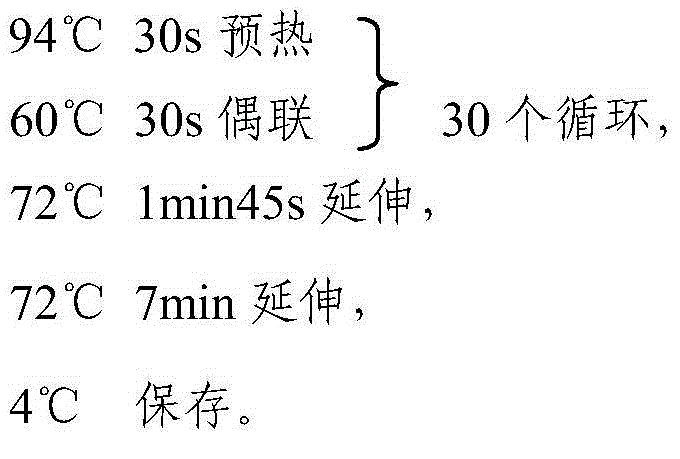
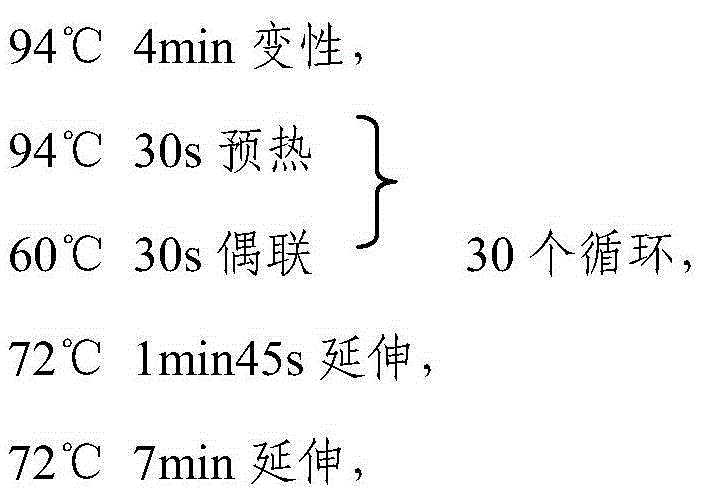
[0037] The primers are designed by the inventors themselves and synthesized by Invitrogen. The PCR reaction system is a 15 μl reaction system, including: 10Xbuffer 1.5 μl; DNTP mixture 1.5 μl (2.5 μm / μl); DNA 2 μl; upstream primer (concentration 10 μm) 0.5 μl; Primer (10pm concentration) 0.5μl; sterile water 8.9μL.
[0038] The PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com