Method and special kit thereof for detecting babesia
A technology for Babesia and Babesia deer, which is applied in the biological field to achieve the effects of improving specificity and improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The design of embodiment 1, primer and probe
[0047] 1. There are three main hypervariable regions in the 18srRNA gene of Babesia, the first is located at the 100-200nt position from the 5' end, the second is located at the 550-660nt position, and the third is located at the 1250- 1300nt position. The difference between different species of Babesia is between 0.0% and 11.2%, and the corresponding positions of the 18srRNA gene of rodent and human origin are close to 30%.
[0048] According to the principle of primer design, a 406bp gene region (SEQ ID No.1) containing the second hypervariable region (550-660 nucleotides from the 5' end) of the 18srRNA gene of Babesia (the sequence was derived from divergence Babesia divergens (Babesia divergens, sequence accession number GU057385 in GenBank) was further analyzed, wherein the bold part is the variation region of different Babesia species.
[0049] SEQ ID No. 1:
[0050] 5’-AATTACCCAATCCTGACACAGGGAGGTAGTGACAAGAAATAACAA...
Embodiment 2
[0059] Embodiment 2, common PCR amplification verification
[0060] 1. Synthesize the fragment shown in SEQIDNo.1 and insert it into the plasmid vector pMD18-T to obtain a recombinant plasmid, which is named pBabesi406. The recombinant plasmid pBabesi406 was sent for sequencing, and the result was correct.
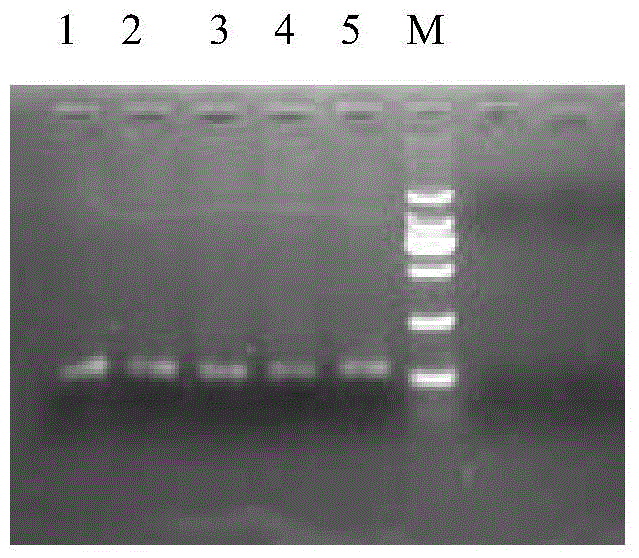
[0061] Two, with the recombinant plasmid pBabesi406 of different gradient dilutions as a template, with SEQIDNo.2 and SEQIDNo.3 (y=t / c, s=g / c, the equal concentration mixture of four kinds of primers) as primers, carry out common PCR amplification Each PCR amplification product was obtained, and the agarose gel electrophoresis results of each PCR amplification product were as follows: figure 1 shown.
[0062] figure 1 Among them, 1-5 are PCR amplification products; M is DNAmarker.
[0063] figure 1 It shows that the target band of about 120bp can be obtained by PCR amplification, and the target band is sequenced, and the result is consistent with the sequence of SEQID...
Embodiment 3
[0064] Embodiment 3, real-time fluorescent quantitative PCR verification
[0065] 1. Insert the DNA fragment shown in SEQIDNo.5 into pMD18-T to obtain a recombinant plasmid, which is named pBabesi116. The pBabesi116 was sent for sequencing, and the result was correct.
[0066] Two, according to the base number (2692+116=2808bp) of the recombinant plasmid pBabesi116, calculate the molecular weight (MW=2808×660=1853280Daltons), that is, 1mol=1.85328×10 6 g.
[0067] The actual nucleic acid concentration of pBabesi116 was measured and repeated three times. The results were 147, 133, and 127 (ng / uL) respectively, and the average value was 135.7 ng / μL, which was used as a standard.
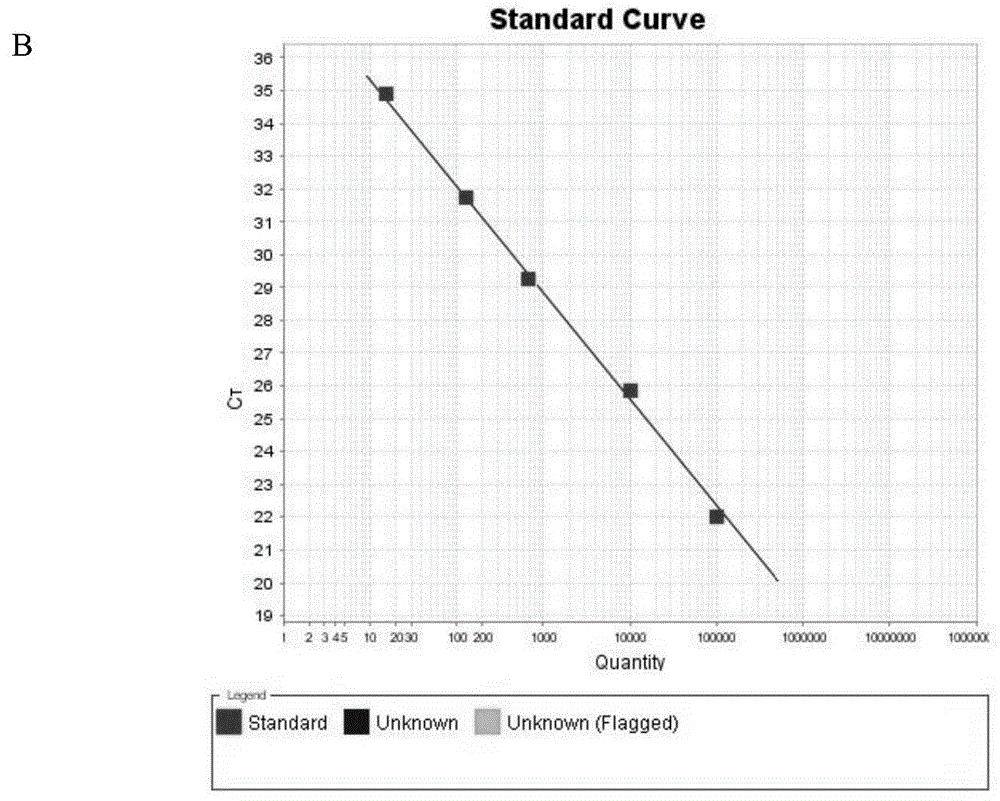
[0068] Calculate standard copy number: (6.02x10 23 copy number / mole)x(1357x10 -10 g / μl) / (1.85328x10 6 g / mol)=4.4x10 10 copies / μL, dilute the standard in different gradients, and calculate the corresponding concentration at the same time, as shown in Table 1.
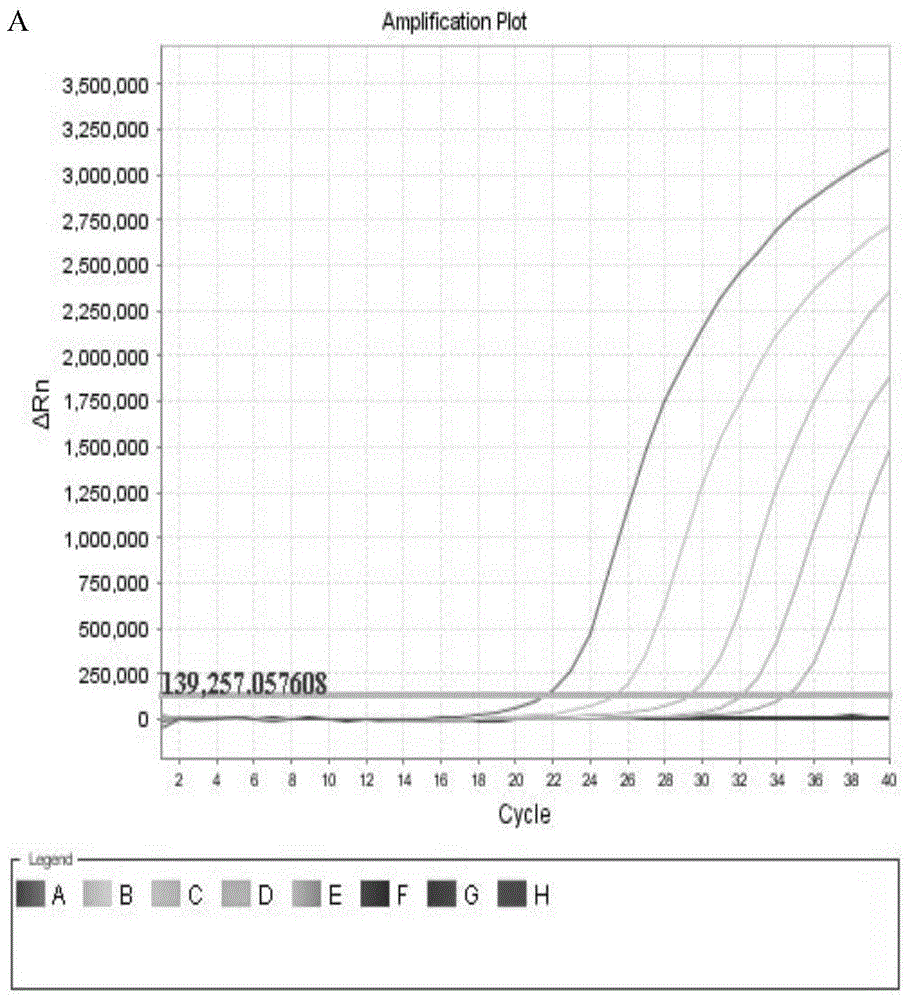
[0069] 3. PCR detection was perform...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com