Method for determining up-regulated genes and down-regulated genes in cotton fiber development process
A development process, cotton fiber technology, applied in the field of plant genes and their encoded proteins, can solve problems such as unclear fiber quality mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0025] According to one embodiment of the present invention, there is provided a method for determining up-regulated or down-regulated genes during cotton fiber development, comprising the following steps:
[0026] Step 1: Obtain the first transcriptome sequencing data and the second transcriptome sequencing data
[0027] Obtain the first transcriptome and the second transcriptome from the samples in the first and second stages of cotton fiber development, respectively, sequence the first and second transcriptomes, and obtain the first transcriptome sequencing data and the second transcriptome The sequencing data, the first transcriptome sequencing data and the second transcriptome sequencing data each comprise a plurality of reads. The samples in the first stage of cotton fiber development and the second stage of cotton fiber development are all derived from cotton ovules.
[0028] In a specific embodiment of the present invention, the first stage of cotton fiber developme...
Embodiment 1
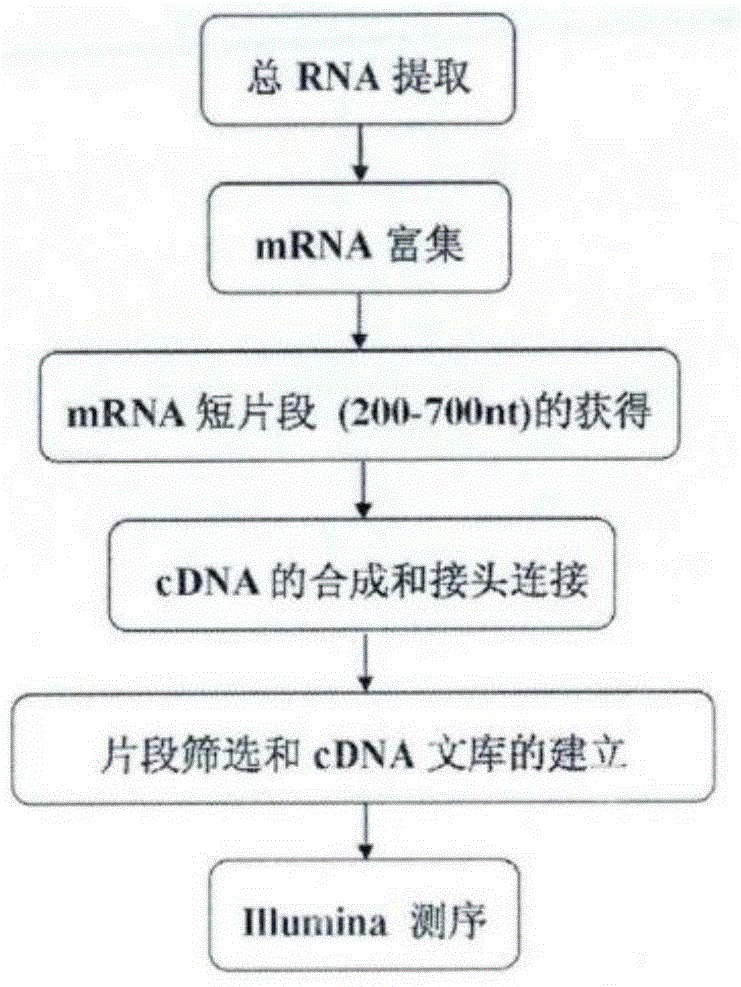
[0053] Example 1: Transcriptome library construction and sequencing
[0054] 1.1 Experimental materials and RNA extraction
[0055] Upland cotton Xinluzhong 36 (Gossypiumhirsutum) Gh36 and Haidao cotton Xinhai 21 (Gossypiumbarbadense) Gb21 were obtained from the Agricultural Science Research Institute of the First Division of Alarnong, Xinjiang. Two varieties of cotton were sown in the field, and the ovules (0DPA) on the day of flowering of Upland cotton and Sea Island cotton were taken respectively. The samples were named xianyang_LZC and xianyang_HZB, and were quickly placed in liquid nitrogen and stored in a -80℃ refrigerator for later use.
[0056] Upland cotton (Gossypiumhirsutum) Gh36 and sea island cotton (Gossypiumbarbadense) Gb21 were the experimental materials (provided by the Agricultural Research Institute of the First Division of Alarnong, Xinjiang). The two strains of cotton were sown in the field, and the fibers of 5, 10, 15, and 25 DPA were picked respectively...
Embodiment 2
[0062] Example 2: Screening of differentially expressed genes
[0063] ——Based on the sequencing data of the same cotton species at different cotton fiber development stages or different cotton species at the same cotton fiber development stage
[0064] 2.1 Transcriptome data statistics and filtering
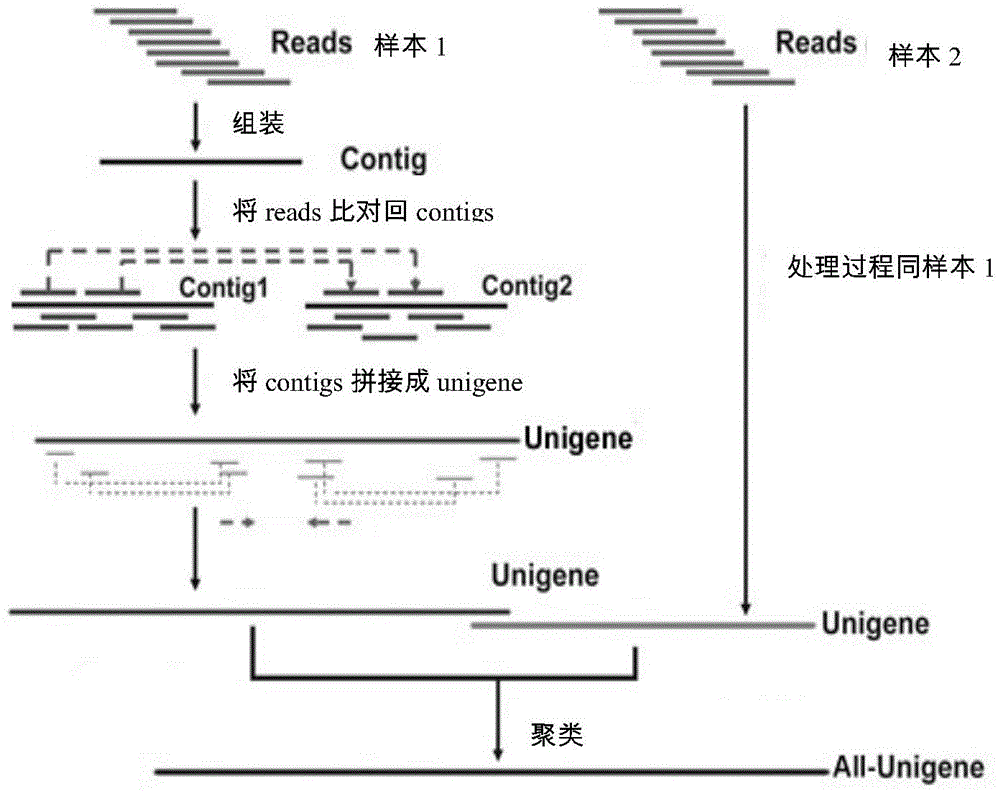
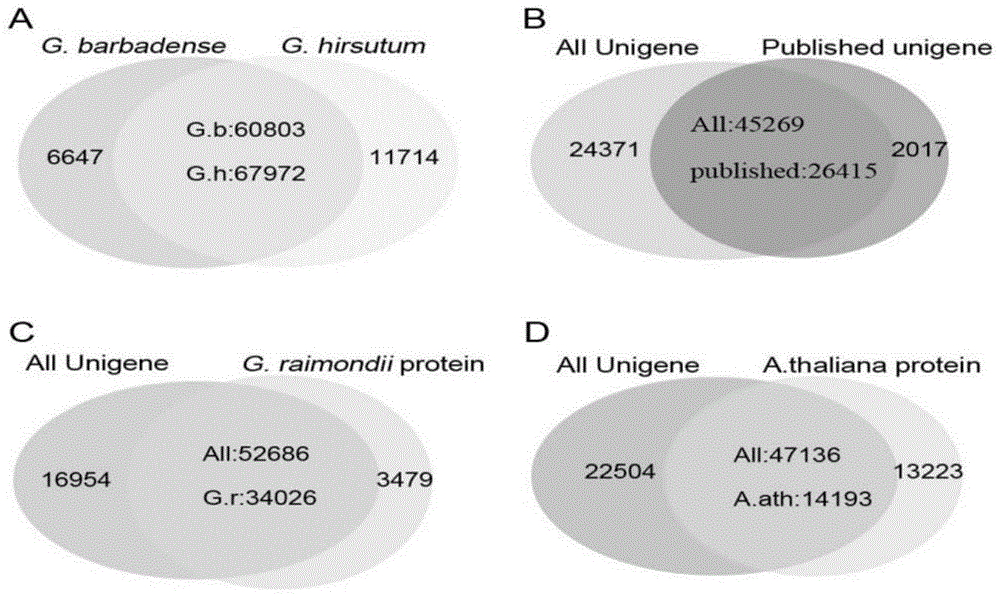
[0065] Filter the raw data (rawreads or rawdata) according to the requirements. Rawdata or rawreads are obtained from the raw image data obtained by base calling (basecalling) transformation and sequencing. Generally, the contamination of the connector, the repetition of the quality value caused by the PCR in the library construction process, and the quality value The very low reads are removed, and clean reads (Cleanreads) are obtained for subsequent analysis, such as obtaining non-redundant gene sequences Unigenes through multiple assemblies, and using bioinformatics for Unigenes to screen differentially expressed genes and / or analyze differentially expressed genes function, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com