Primer pair and method for identifying hericium erinaceus houjie No.2
A technology of Hericium erinaceus and primer pairs is applied in the directions of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., which can solve the problems of complex PCR amplification patterns, unsuitable for variety identification, and low specificity, etc., and achieve identification operations. Convenience, short detection time, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
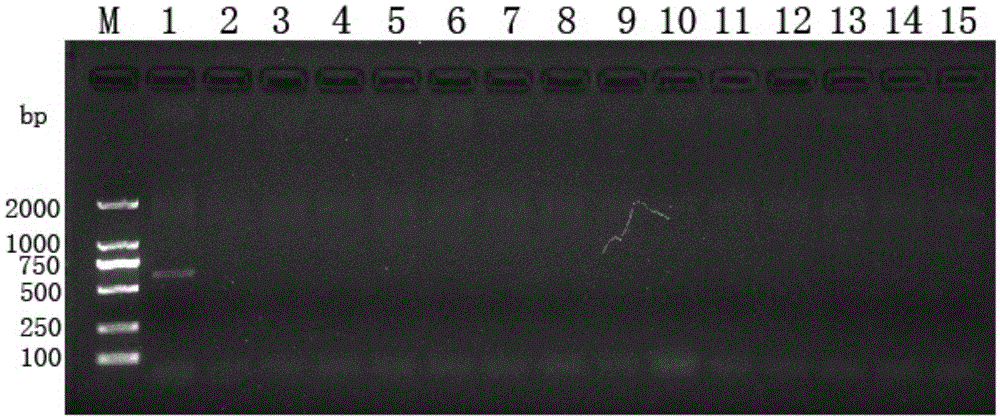
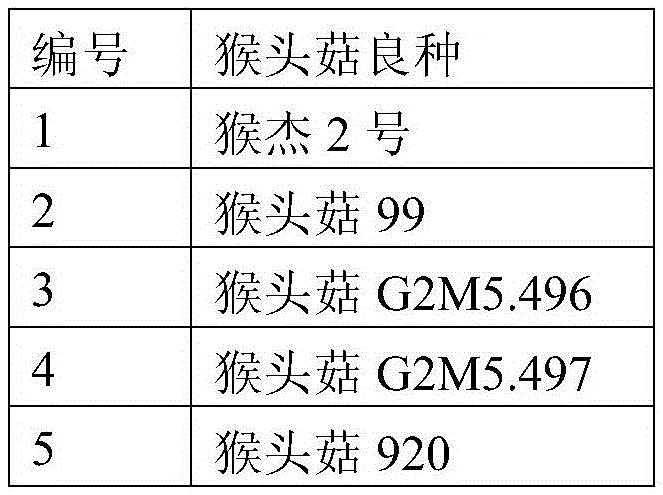
[0031] An exploratory study was carried out on the molecular identification of 15 main species of Hericium erinaceus using SRAP molecular marker technology. Extract the hyphae genome DNA of each Hericium erinaceus species as a template, carry out PCR amplification with a large number of SRAP random primer pairs, and detect the amplified products by agarose gel electrophoresis, and finally screen to obtain differential bands (only Hericium erinaceus L. No. 2 has a pair of primers but the other varieties do not). The differential band was recovered by gel cutting, then connected to the cloning vector and sent for sequencing. The size of the obtained DNA fragment was 597bp, and its base sequence was shown in SEQIDNo.1. This DNA fragment was the specific DNA of Houjie 2 fragment.
[0032] A primer pair was designed for the specific DNA fragment of Houjie 2 obtained, and the sequence of the primer pair is:
[0033] Upstream primer: 5′-CAAACCGGTGTCAGGGAACTGC-3′
[0034] Downstrea...
Embodiment 2
[0038] (1) Extraction of Hericium erinaceus Genomic DNA
[0039] Take 0.03g of Hericium erinaceus mycelium to be tested, add liquid nitrogen to grind thoroughly, use a new rapid plant genome DNA extraction kit (DP3111, BioTeke, Beijing BioTeke Biotechnology Co., Ltd.) to extract genomic DNA, and obtain improved varieties of Hericium erinaceus Genomic DNA crude extract. The integrity, purity and concentration of the obtained crude genomic DNA were detected by agarose gel electrophoresis and a DNA / RNA UV spectrophotometer (Nanodrop Technologies, USA). DNA samples with OD260 / OD280>1.8 were used for subsequent PCR amplification. DNA extracts were stored in a -20°C refrigerator for later use.
[0040] (2) PCR amplification
[0041] Composition of PCR reaction solution: 2×PowerTaqPCRMasterMix 10μL, 10μM upstream and downstream specific primer pairs 1.5μL each, 20ng / μL template DNA 3μL, ddH 2 04 μL.
[0042] The amplification reaction was carried out on a LifeECO amplification ins...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com