A method for finding zinc finger protein target sites in the genome
A technology of zinc finger protein and target site, which is applied in the field of genetic engineering, can solve problems such as spending a lot of time, and achieve the effects of improving efficiency, saving costs, and simplifying experimental procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Searching for zinc finger protein target sites in the silkworm partial genome sequence nscaf2681
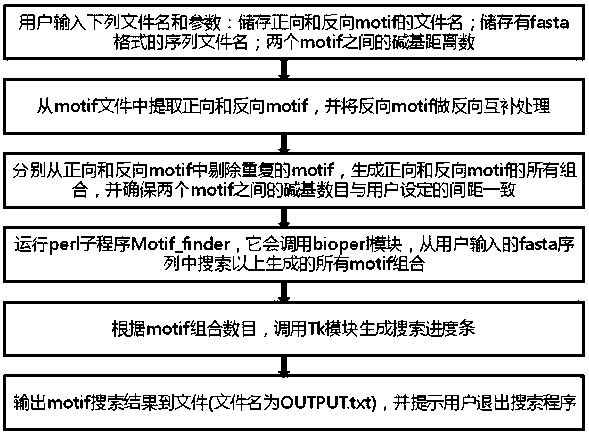
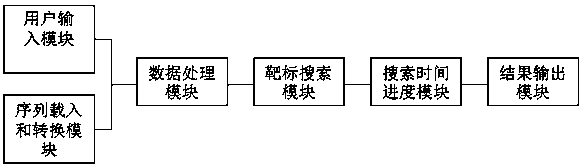
[0040] 1. The search method is as follows: first, all 9 nucleotide sequences that can bind to zinc finger proteins and have a binding rate greater than 2 (that is, all zinc finger protein genes that can bind to 9 nucleotide sequences and have a binding rate greater than 2) Build a library; then design a software program, use the program to search for sites that match the 9 nucleotide sequences in the library described in step S1 at intervals in the double strand of the target genome, and obtain the corresponding Zinc finger protein target sites.
[0041]2. The specific operation method is as follows (operating system requirements: Windows system):
[0042] (1) Name the genome sequence nscaf2681 as seqnscaf2681.txt;
[0043] (2) Name the file name of the library consisting of all 9 nucleotide sequences whose binding rate to the zinc finger protein is greater tha...
Embodiment 2
[0057] Example 2 Searching for zinc finger protein target sites in the silkworm partial genome sequence nscaf1705
[0058] The basic requirements and process are the same as those in Embodiment 1, and the system requirements: Windows system.
[0059] The specific operation method is as follows:
[0060] (1) Name the genome sequence nscaf2681 as seqnscaf1705.txt
[0061] (2) Name the file name of the library composed of all 9 nucleotide sequences whose binding rate to zinc finger protein is greater than 2 motifs.txt
[0062] (3) Set the number of base distances between the forward and reverse motifs of the zinc finger protein target site to be 7;
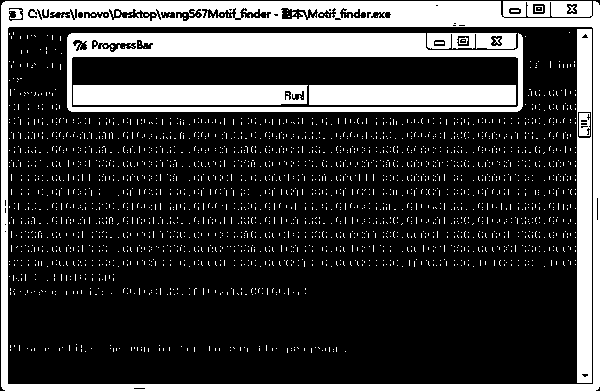
[0063] (4) The program runs the command Mptif_finder.exe
[0064] The prompt is as follows: Enter your motif file name and fasta file name that contains all the primers and target sequences, specifically:
[0065] Type motifs.txt seqnscaf1705.txt, press enter, click the Run button, and the program starts to run;
[0066] (5) Pre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com