Three-generation PacBio sequencing data-based hole filling method
A technology for sequencing data and filling holes, applied in the field of biological information, can solve the problems of slow comparison speed, waste, and long scaffolding time, and achieve the effects of improving accuracy, saving memory, and saving comparison time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
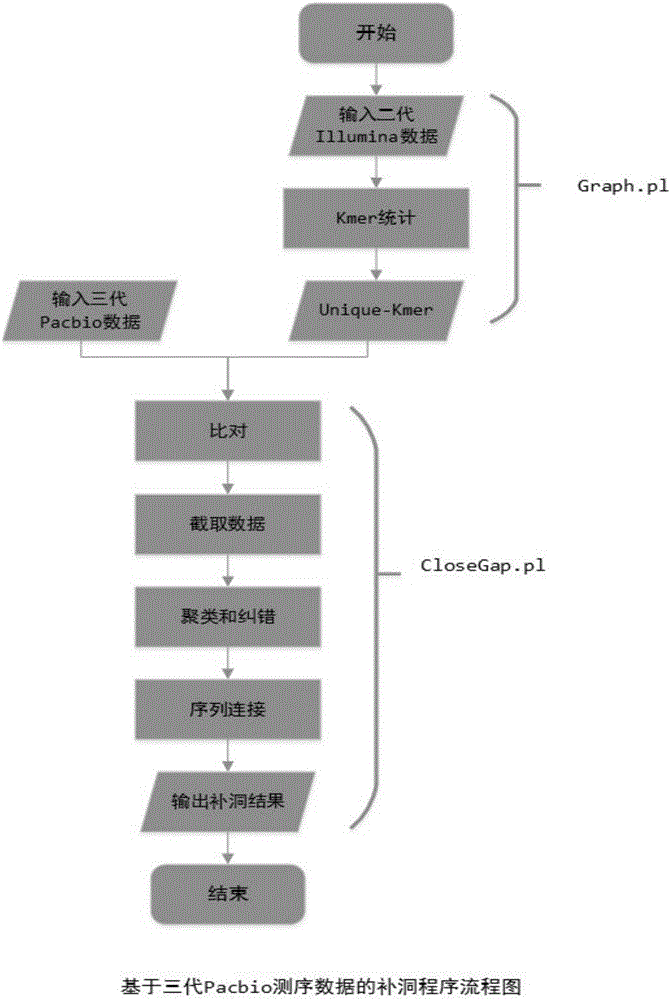
[0041] 1. Extract the unique-kmer from the contig. In step (1), use the Jellyfish software to perform k-mer statistics on the second-generation Illumina sequencing data, and use the k-mer that appears once as the unique-kmer. For k≤17, use a size It is stored as a 2G bit file (*.bit file), and for the case of k>17, the unique-kmer is stored in the (*.h5) file in the GATB open source package. Wherein, breaking all the data into fragments of length k is called k-mer, and the second-generation Illumina sequencing data refers to the next-generation sequencing data obtained through the sequencer of Illumina Company.
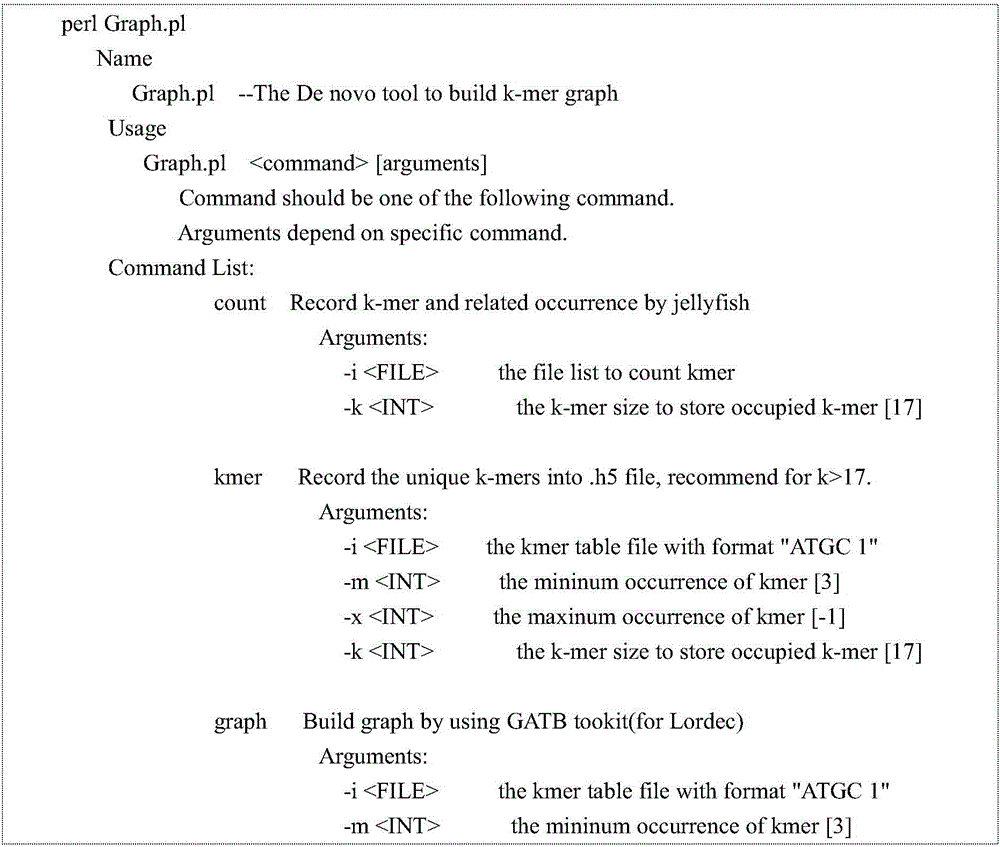
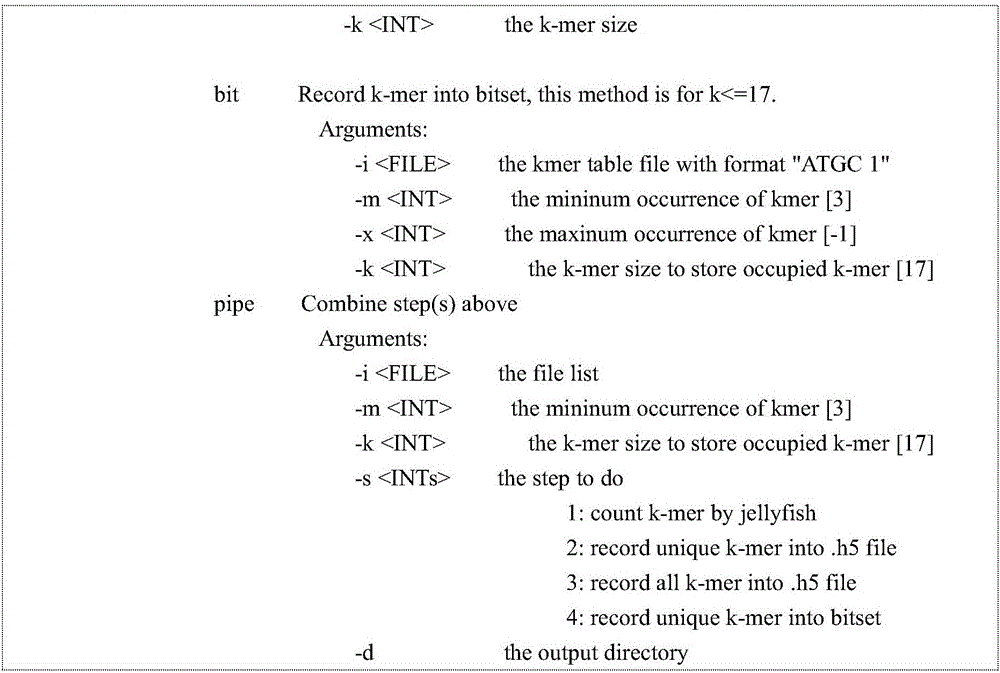
[0042] The program was written according to the above method, and the usage is as follows:
[0043]
[0044]
[0045] Put the contig path into a file file.lst
[0046]
[0047] Then run the program to get unique-kmer:
[0048]
[0049] Because k=17 is selected, the result is stored in the bit file: k17.bit
[0050] 2. Use unique-kmer as the seed for com...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com