Serum miRNA marker related to esophageal squamous carcinoma assisted diagnosis and application of serum miRNA marker
A technology for esophageal squamous cell carcinoma and auxiliary diagnosis, which is applied in the fields of genetic engineering and oncology to achieve the effects of good stability, easy access, and minimally invasive access.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
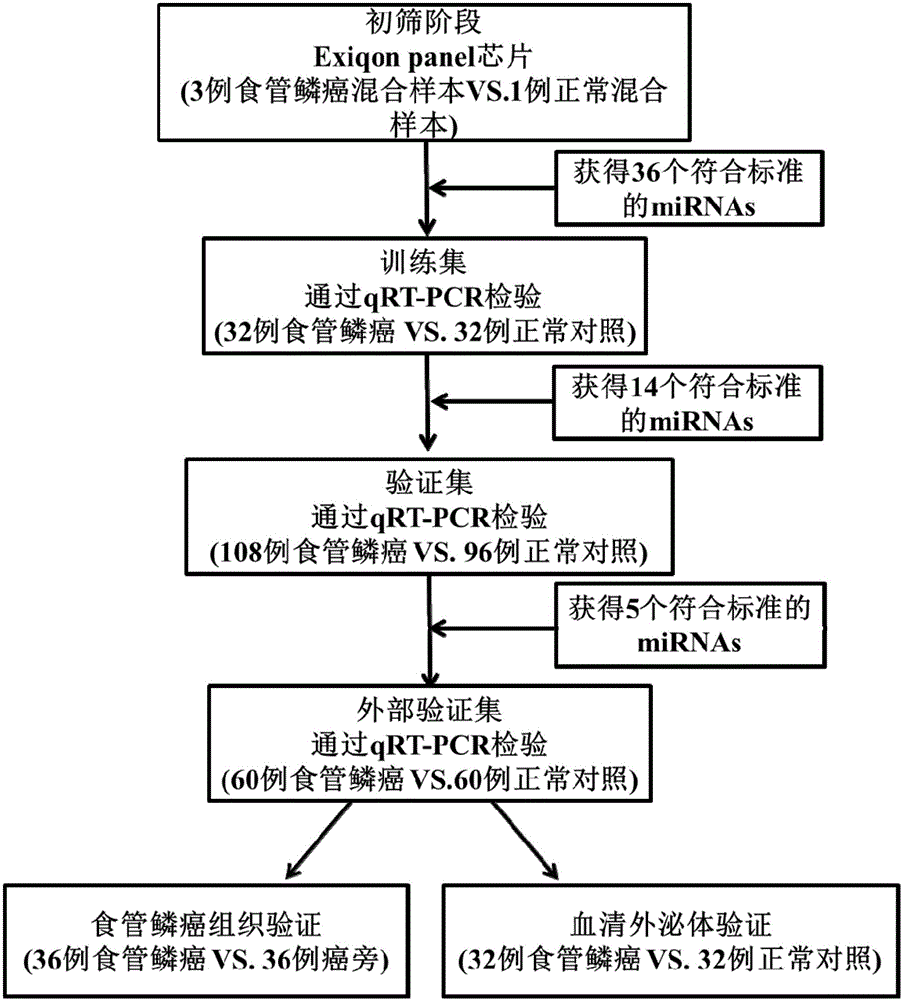
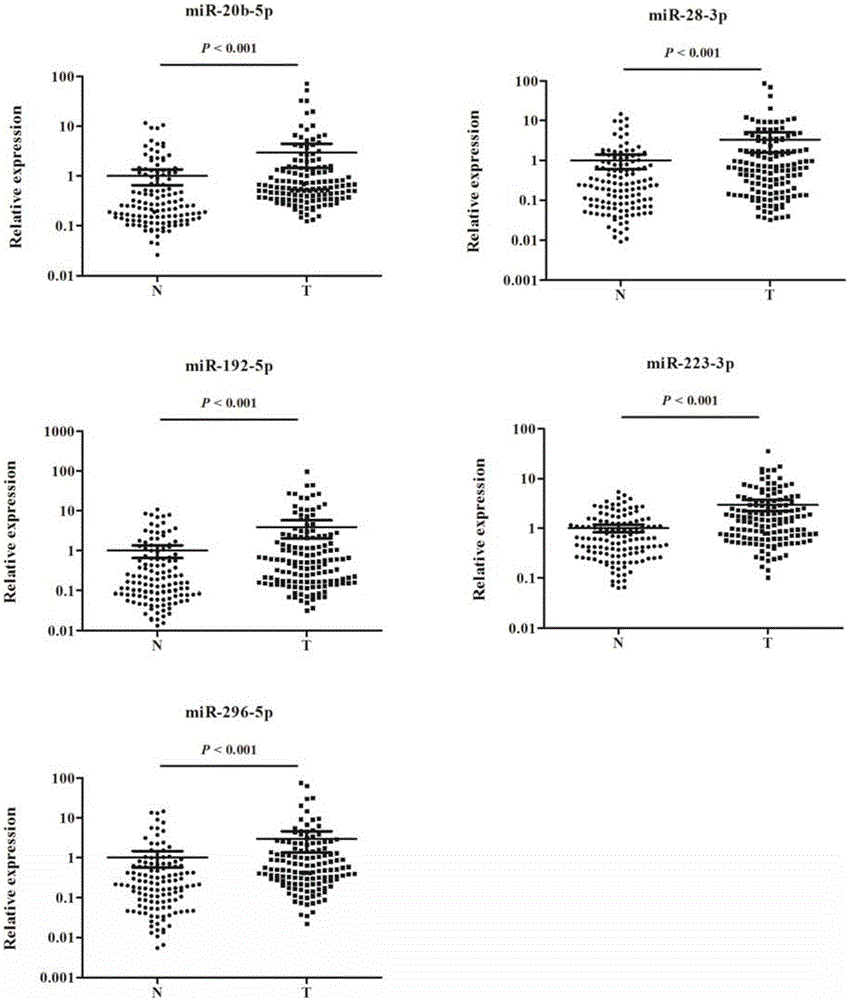
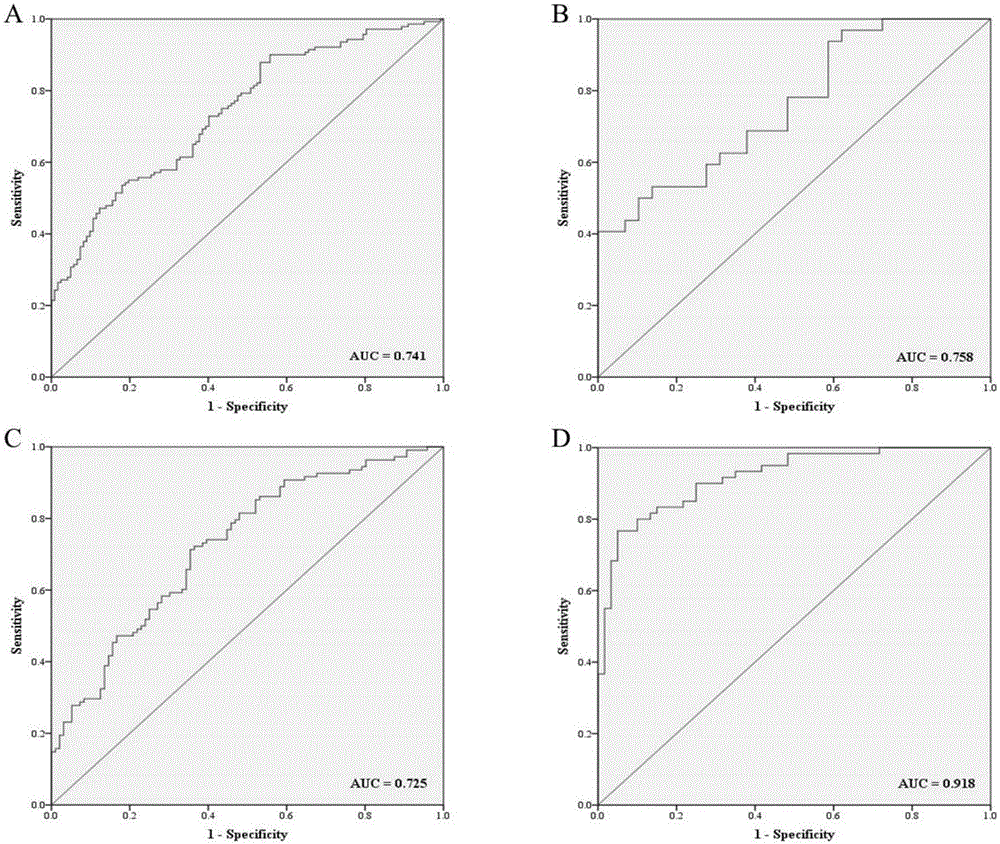
[0036] The inventor collected a large number of venous serum samples from the First Affiliated Hospital of Nanjing Medical University and the First People's Hospital of Changzhou from 2013 to 2015, and selected The samples of 200 cases of esophageal squamous cell carcinoma and 188 cases of normal controls were used as the experimental samples for the initial screening of Exiqon miRNA qPCR panel chip and subsequent series of qRT-PCR verification. At the same time, 36 cases of esophageal squamous cell carcinoma tissues and 36 cases of paracancerous tissues were retained. The selected patients' serum samples were all from patients who had not undergone surgery, radiotherapy and chemotherapy intervention and were pathologically confirmed as esophageal squamous cell carcinoma. The demographic data and clinical data of these samples were collected systematically.
[0037] Refer to the flowchart ( figure 1 ), randomly selected 30 esophageal squamous cell carcinoma samples and 10 no...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com