Long non-coding RNA and application thereof
A long-chain non-coding, non-coding technology, applied in the direction of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem of not finding colorectal cancer IncRNA and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
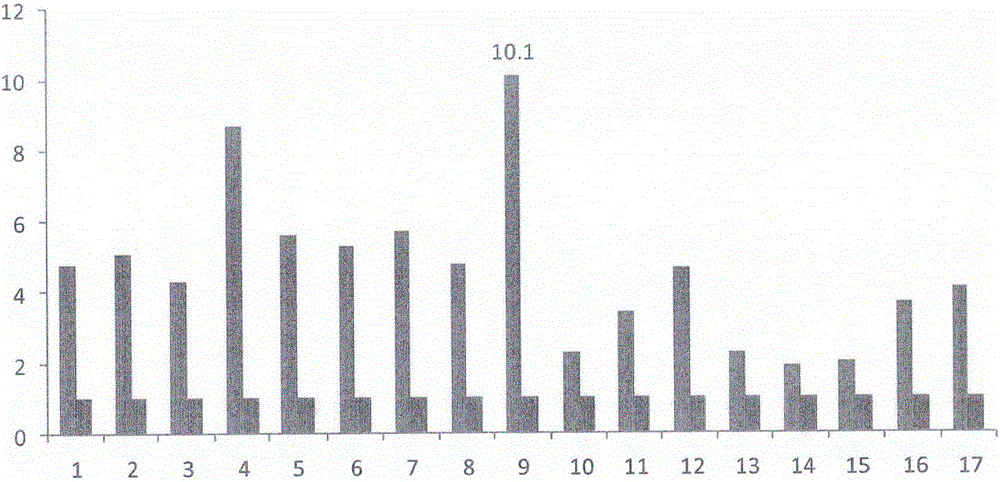
[0020] Example 1 Detection of TUC338 Expression in Colorectal Cancer Tissue Samples
[0021] 1) Fresh cancer tissues and their corresponding paracancerous tissues (beyond the resection margin of 3cm) were selected from 17 patients with colorectal cancer who were diagnosed with colon cancer or rectal cancer by surgery. Chemotherapy, radiotherapy and other anti-tumor treatments are performed to rule out other systemic diseases and the possibility of metastatic cancer, and the functions of all important organs are within the normal range.
[0022] 2), the synthesis of primers: query in GenBank Blast, find the sequence (SEQID NO: 1) of the target gene TUC338mRNA, apply the express 2.0 software of Primer to design specific primer probes on the CDS region, and the primer probes are synthesized by (company ) synthesis, the primer sequence is as follows:
[0023] Forward (5'-3): GCCCCACCCCCTTCATTCTTA (SEQ ID NO: 2)
[0024] Reverse (5'-3'): CAGACACAATTTGAAACGAGCTG (SEQ ID NO: 3);
...
Embodiment 2
[0057] Embodiment 2 Design and carrier preparation of interfering RNA sequence
[0058] 1) Design interference fragments and negative control sequences according to the sequence information of TUC338, wherein the interference sequences are:
[0059] siRNA-TUC338:
[0060] Sense Seq: 5'-UACAGCACAGGCAGCGACTT-3'; (SEQ ID NO: 2)
[0061] Antisense Seq: 5′-GUCGCUGCCUGUGCUGUATT-3′;
[0062] Negative Control siRNA:
[0063] Sense Seq: 5'-UUGUGCGAAGCUGAUGCUTT-3'; (SEQ ID NO: 7)
[0064] AntiSense Seq 5'-AGCAUCAGCUUCGCACAATT-3'.
[0065] 2) Vector construction
[0066] The above-mentioned fragments are chemically synthesized, and after annealing, they are ligated into the vector pLVX-shRNA1 by enzyme digestion. The method includes:
[0067] a) Annealing of oligonucleotides
[0068] Dilute the synthesized deoxynucleotides to 1 μg / μL, take 5 μL each of the sense strand and the antisense strand and anneal to form a double strand. The reaction system is as follows:
[0069] Sense s...
Embodiment 3
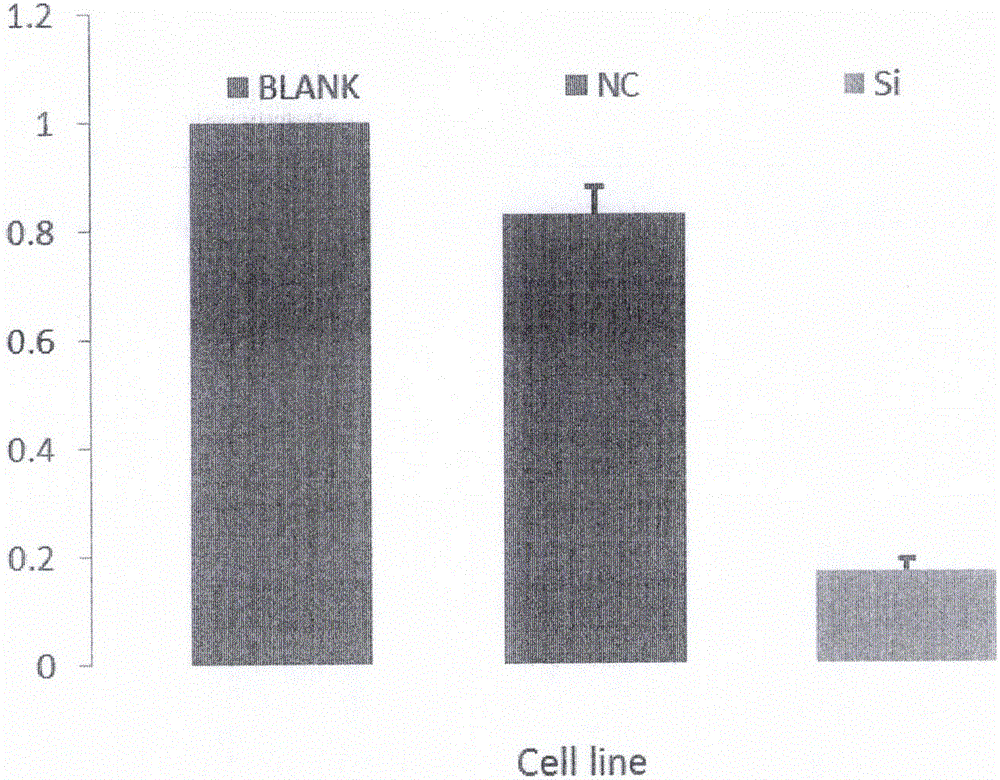
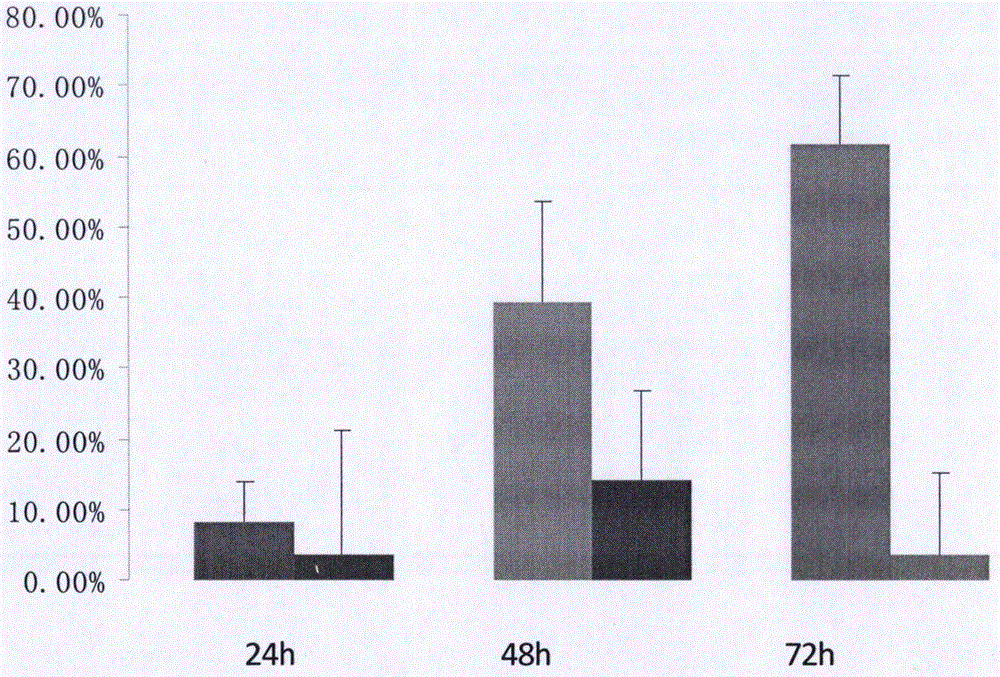
[0089] Example 3 Interference Effect Test of Interfering RNA Carrier
[0090] HT29 cells were used as recipient cells, which were resuscitated before transfection and subcultured; experimental groups were set up, namely interference group (Si), negative control group (NC) and blank control group (Blank).
[0091] 1) Cell transfection
[0092] a) 24 hours before transfection, the cells were replaced with a medium that did not contain any antibiotics;
[0093] b) the cell confluency reaches 70%;
[0094] c) Add 20 μL of the ligation product to 400 μL of serum-free medium, add 5 μL of Lipo2000 transfection reagent, mix well at room temperature, and let stand for 20 minutes;
[0095] d) Set up three replicate wells for each group, add 400 μL of the transfection reagent prepared in the above steps to each well, and place in 5% CO 2 , cultured in a 37°C incubator for 7-8h, and then replaced with normal culture medium.
[0096] e) 24-48 hours after transfection, detect the RNA ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com