Method for distinguishing mating types of four varieties in black morchella group and primer pair
A technology for mating type and Morchella, applied in the field of biology, can solve problems such as loss of mating type, and achieve the effects of simple method and stable and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Identification of the mating type of Morchella edulis YPL2 monoascospore strain
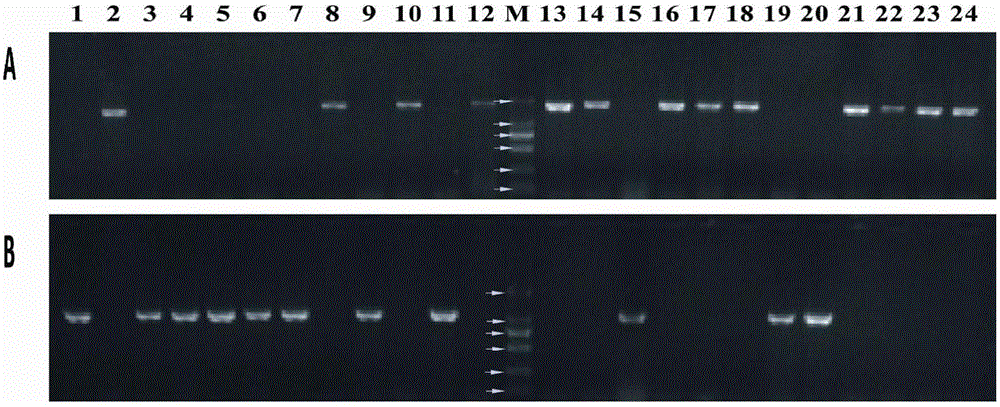
[0045] The monoascospore strains YPL2-1—YPL2-24 of Morchella chinensis were inoculated on PDA medium, cultivated at 23°C for 10 days, picked appropriate amount of mycelia, extracted DNA by CTAB method, and took 3 μL in 1% (W / V ) agarose gel electrophoresis to detect DNA quality and concentration.
[0046] Using the DNA of YPL2-1—YPL2-24 as a template, PCR amplification was performed on P8-5F / P8-5R with primers: 25 µL amplification system included 2×PCRmix (TSINGKE Bio Inc) 12.5 µL, 10 µM / L 1 µL each of P8-5F and P8-5R primers, 1 µL of DNA template, ddH 2 O 9.5 µL. The PCR amplification program was: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, extension at 72°C for 90 sec, 30 cycles; extension at 72°C for 10 min.
[0047] Using the DNA of YPL2-1—YPL2-24 as a template, PCR amplification was performed on P10-1F / P10-2R with pri...
Embodiment 2
[0049] Example 2 Mating Type Identification of Morchella HL1 Monoascospore Strain
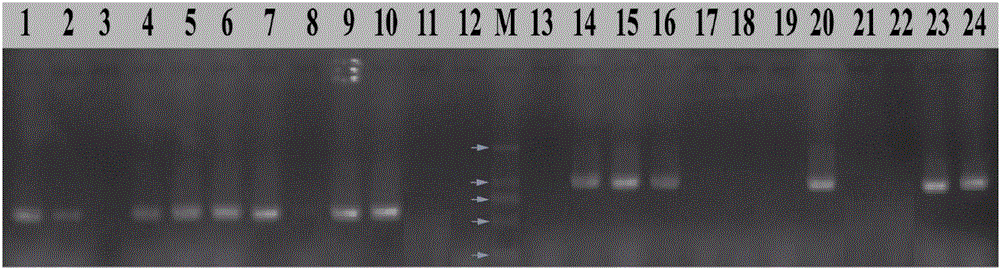
[0050] The monoascospore strain HL1-1—HL1-12 of Morchella liumei was inoculated on PDA medium, cultured at 23°C for 10 days, picked appropriate amount of mycelium, extracted DNA by CTAB method, and took 3 µL on 1% agarose gel DNA quality and concentration were detected by electrophoresis.
[0051] Using the DNA of HL1-1—HL1-12 as a template, PCR amplification was performed on P7-2F / P7-2R with primers. The 25 µL amplification system included 2×PCRmix (TSINGKE Bio Inc) 12.5 µL, 10 µM / L 1 µL each of P7-2F and P7-2R primers, 1 µL of DNA template, ddH 2 O 9.5 µL. The PCR amplification program was as follows: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 sec, annealing at 62°C for 30 sec, extension at 72°C for 40 sec, 30 cycles; extension at 72°C for 10 min.
[0052] Using the DNA of HL1-1—HL1-12 as a template, PCR amplification was performed on P10-2F / P10-3R with primers. The 25...
Embodiment 3
[0054] Example 3 Identification of the mating type of the morel 2688 monoascospore strain
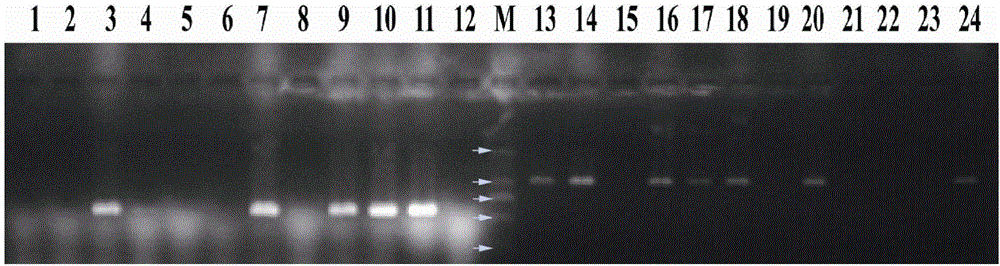
[0055] The monoascospore strains 23-S1—23-S12 of Morchella soliformis were inoculated on PDA medium, cultured at 23°C for 10 days, an appropriate amount of mycelium was picked, DNA was extracted by CTAB method, and 3 µL was electrophoresed on 1% agarose gel Check DNA quality and concentration.
[0056] Use the primer pairs P7-2F / P7-2R and P10-2F / P10-3R to detect the mating type, and the operation steps are the same as in Example 2.
[0057] All amplified products were detected by electrophoresis on 1.2% agarose gel, the results are shown in image 3 , the strains 23-S3, 23-S7, 23-S9, 23-S10, and 23-S11 only amplified a band of about 0.6kb by the primer pair P7-2F / P7-2R, which was the mating type of MAT1-1; the strain 23- For S1, 23-S2, 23-S4, 23-S15, 23-S6, 23-S8, and 23-S12, only the primer pair P10-2F / P10-3R amplifies a band of about 1.0 kb, which is MAT1-2 mating type .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com