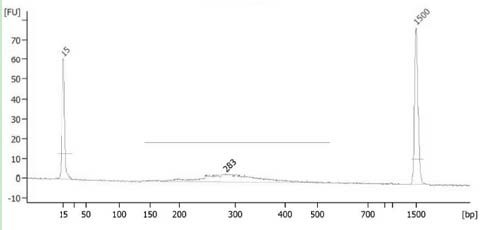
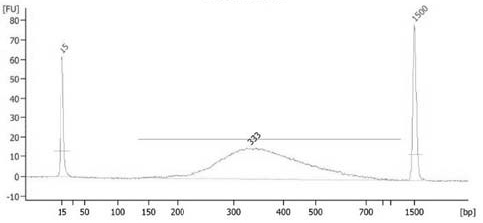
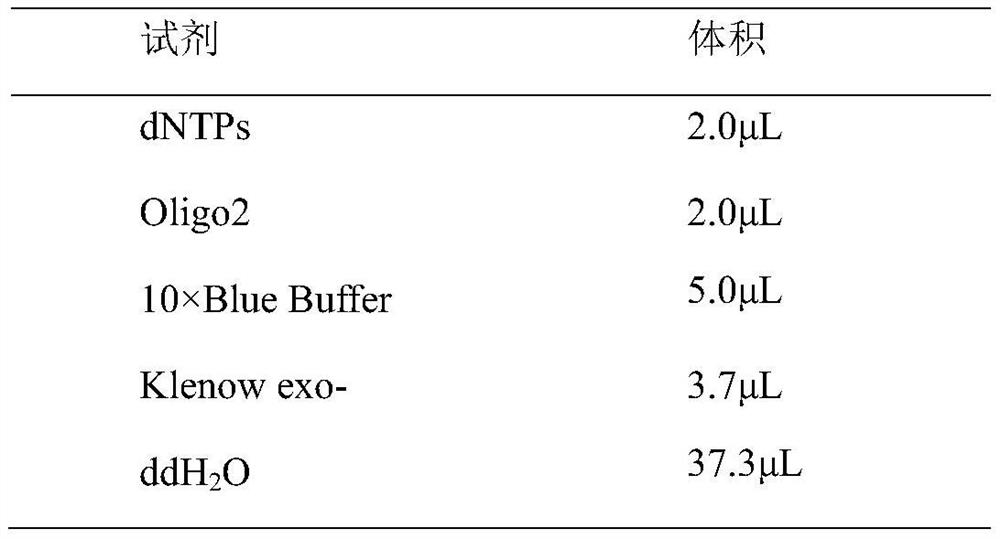
A Library Construction Method for Single-Cell Whole-Genome Bibisulfite Sequencing
A bisulfite and construction method technology, applied in the field of library construction of single-cell whole-genome bisulfite sequencing, can solve the problems of low data comparison rate, etc. The effect of improving the data comparison rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The present invention will be described more specifically by way of examples below. It should be understood that the embodiments described here are used to explain the present invention, not to limit the present invention.
[0070] 1. Collection of Single Cells
[0071] Single cells were collected in a volume within 0.5 μL into 0.2 mL thin-walled PCR tubes containing 1 μL of calcium and magnesium-free PBS buffer. The collected single cells are named according to the following format.
[0072]
[0074] Prepare Lysis Buffer 10mM Tris-Cl (pH 7.4), 2% SDS. The single-cell sample in a 0.2mL centrifuge tube has a volume of about 1μL. No need to transfer the tube, directly add 11.5μL Lysis Buffer and 0.5μL proteinase K to the tube, vortex slightly to mix, and then centrifuge. React for 1 hour at 37°C on a PCR instrument, and add ddH after the reaction 2 O to a total volume of 20 μL.
[0075] 3. Bisulfite treatment
[0076] The sample obtained i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com