A method and device for simultaneously detecting methylation level, genome variation and insertion fragment
An insert and methylation technology, applied in the field of bioinformatics, can solve the problems of DNA damage, inability to detect DNA sequencing data, and inability to perform genomic variation and insert analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1: the application example of the inventive method
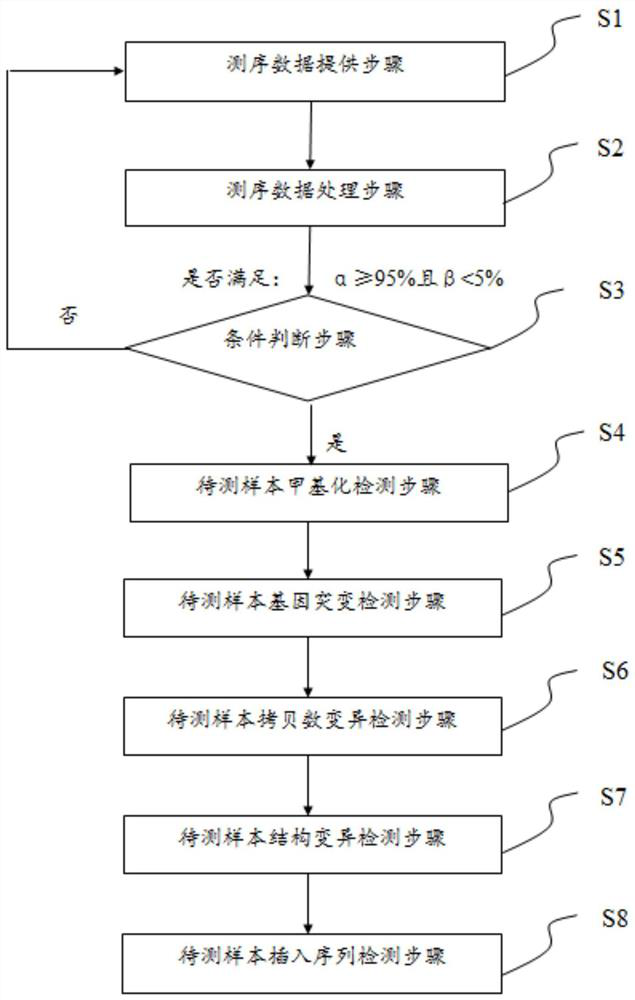
[0086] S1 step:
[0087] Take 100ng of human blood cfDNA, 0.2ng of methylated pUC19 DNA (methylated ginseng DNA), unmethylated lambda DNA (methylated yin ginseng DNA) and mix them for interruption. Bisulfate sequencing. The construction of the sequencing library refers to Example 4 of the Chinese patent application whose application number is CN201911159400 and the title of the invention is "whole genome methylated non-bisulfite sequencing library and construction" of the applicant of the present invention, and the sequencing platform adopts Gene+Seq platform. After sequencing, get off-machine data L1_R1.fq.gz, L1_R2.fq.gz, L1_R1.clean.fq.gz, L2_R2.clean.fq.gz.
[0088] S2 step:
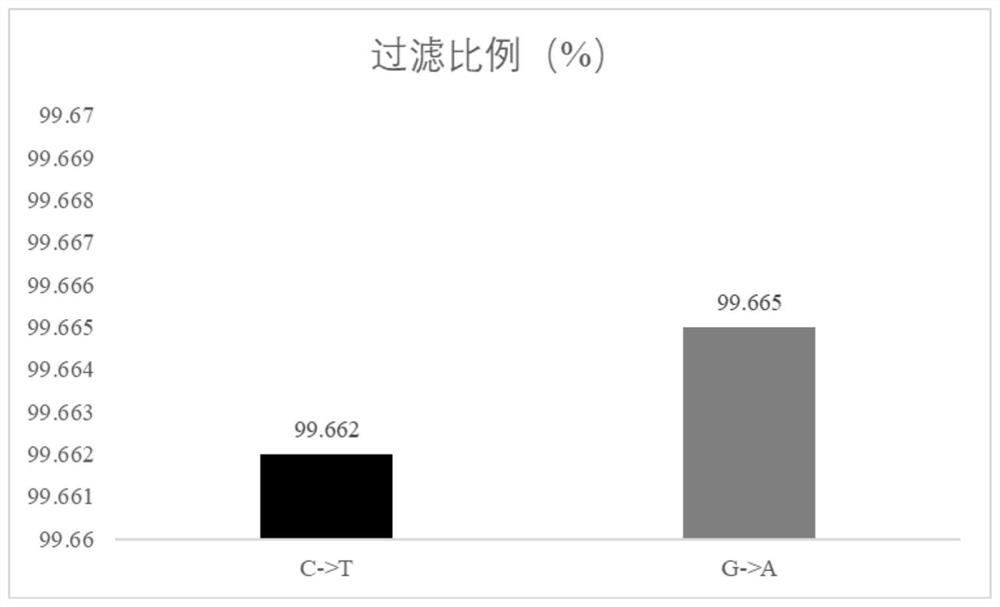
[0089] Use the command fastp-i R1.fq.gz-I R2.fq.gz-o R1.clean.fq.gz-O R2.clean.fq.gz to remove joints and filter the quality of the off-machine data to obtain the filtered Sequence data, as shown in the table below:
[0090]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com