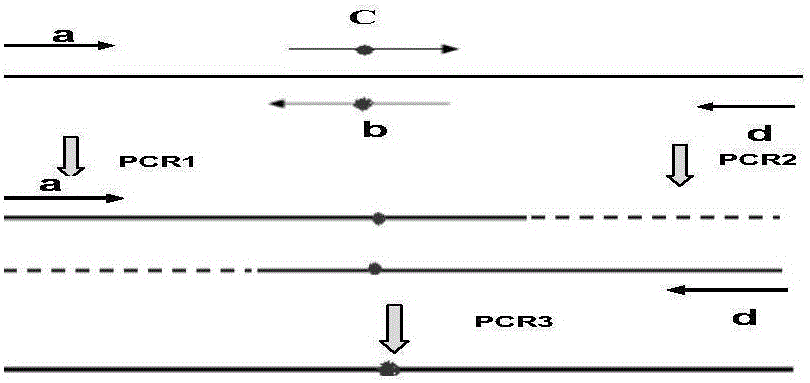
Detection method of gene hot-spot mutation
A gene and mutation gene technology, which is applied in the detection of gene hotspot mutations, genetic engineering and medical diagnosis, can solve the problems that bacteria cannot complete α complementation, cannot form β-galactosidase, destroy the reading frame, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] 1. KRAS gene codon 12 hotspot mutation and corresponding wild sequence plasmid construction
[0066] 1.1 The gene sequence containing the KRAS gene hotspot mutation is shown in Seq No1.
[0067]Primer a TAGCCAAGCTTTGACTGCATACAAACTTGT, as shown in Seq No 2;
[0068] Primer d CGTCAGGATCCTTGGACCATATTCGTCCACA, shown in Seq No3.
[0069] Using the human blood genome as a template, PCR was performed with primers a and d to construct a wild template.
[0070]
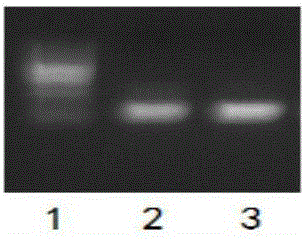
[0071] The above system was reacted on the PCR instrument according to the following procedure: pre-denaturation at 95°C for 5 minutes, followed by a three-step cycle (95°C-15s, 58°C-15s, 72°C-15s, a total of 30 cycles), and finally 72°C-1min. Take 5 μL of the reaction solution for electrophoresis on agarose gel, such as figure 2 Shown: Lane 1 is a 100bp DNA marker; lanes 2 and 3 are wild-type PCR products.
[0072] The PCR product was purified, then connected to the PMD-19T plasmid, and finally transformed into...
Embodiment 2
[0125] 1. The 13th codon hotspot mutation of the KRAS gene and the corresponding wild sequence plasmid construction are the same as above, and the sequences of the selected intermediate primers primers b' and c' are as follows:
[0126] b': ACGTCACCAGCTCCAACTA, as shown in Seq No.8;
[0127] c':TTGGAGCTGGTGACGTAGG, as shown in Seq No.9.
[0128] 2. PCR amplification of the wild and gene fragments containing the 13th codon mutation
[0129] 2.1 Primer g: AGACATCGTAGGTAGTGACATAGCCAAGCTTTGACTGCATAC, as shown in Seq No.10;
[0130] Primer h: AAGGGATCCTTGGACCATATTCGTCCACAAAATGATTCTGAATTAGCTGTATCGTCAAGGCACTCTTGCCTAGG, as shown in Seq No.11.
[0131] The PCR amplification system is as follows:
[0132] -
[0133]
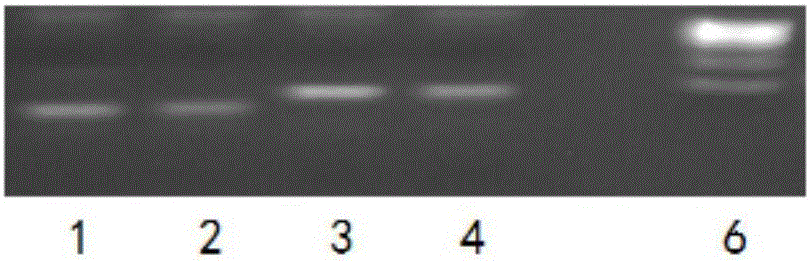
[0134]The above system was reacted in the PCR instrument according to the following procedure: pre-denaturation at 95°C for 5 minutes, followed by three-step cycles (30 cycles of 95°C-15s, 58°C-15s, and 72°C-15s), and finally 72°C-1min.
[0135] The templates were ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com