Site-specific insertional inactivation method and application mediated by agrobacterium tumefaciens and CRISPR/Cas9
A technology of Agrobacterium tumefaciens and genes, applied in biochemical equipment and methods, viruses/bacteriophages, DNA/RNA fragments, etc., can solve problems such as inability to insert precisely at specific sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
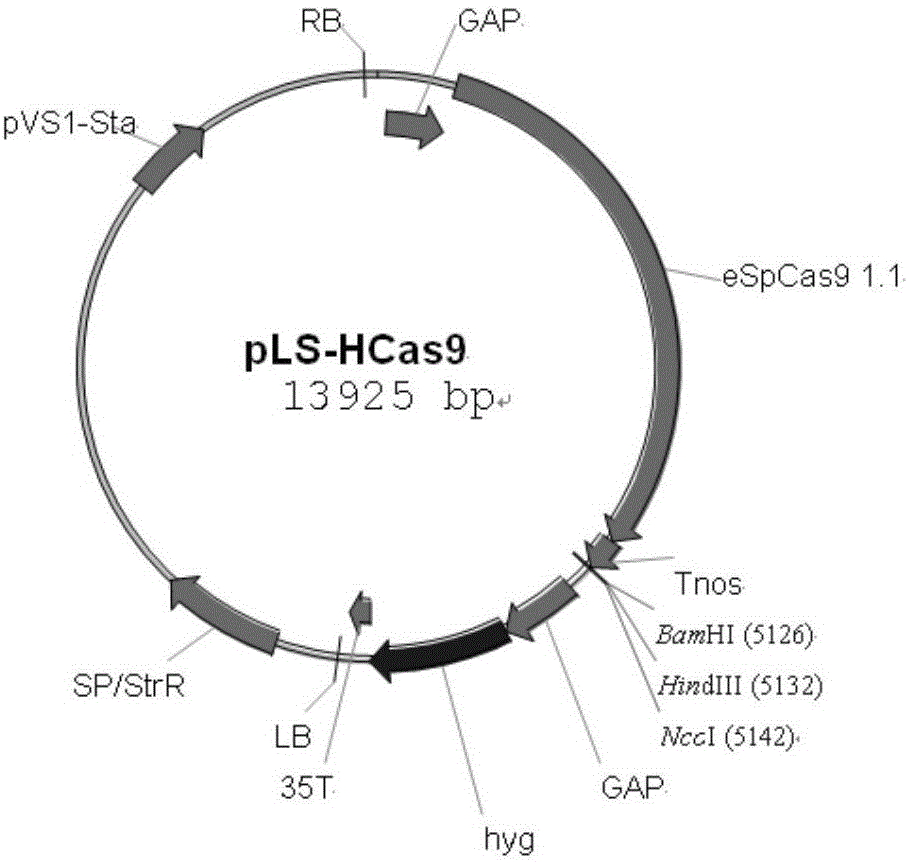
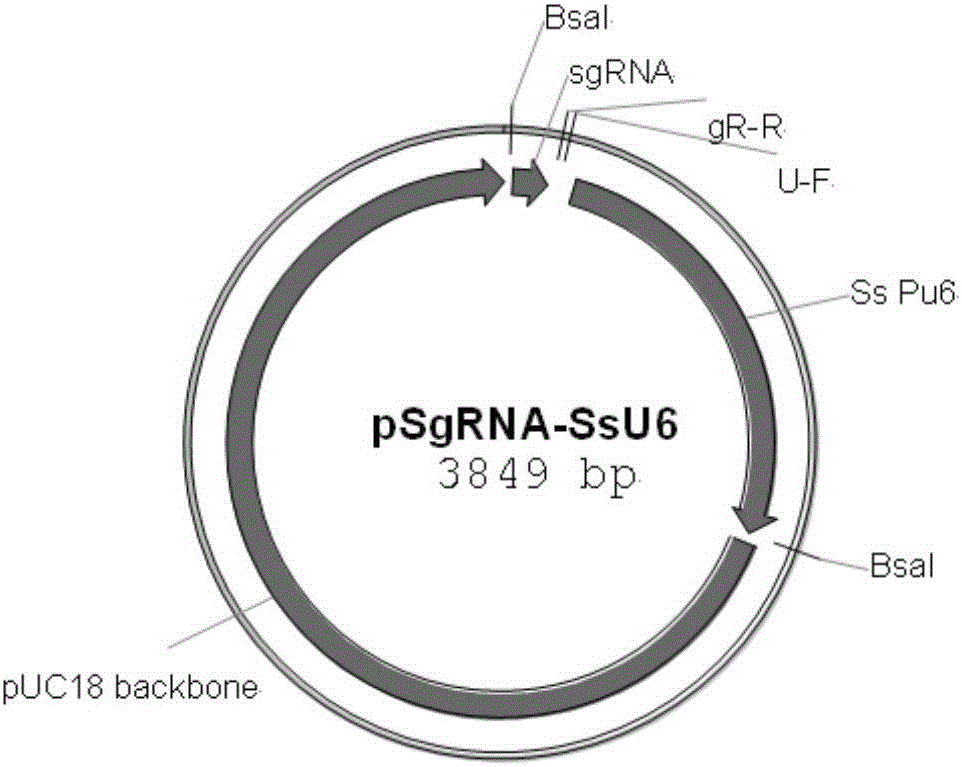
[0025]The Agrobacterium tumefaciens and CRISPR-Cas9-mediated gene site-directed insertion inactivation method of the present invention is through the endogenous U6 gene promoter (Ss PU6, the base sequence of SEQ.ID.No.17; Ss U6RNA, the base sequence of SEQ.ID.No.18) was fused with the target sequence of the inserted site and the sgRNA sequence by Overlapping PCR, and recombined with the endogenous gapd gene of Ustilago sativa by In-fusion technology In the binary vector of the Cas9 gene and hygromycin resistance gene driven by the subunit; then Agrobacterium tumefaciens carrying the binary vector mediated the transformation of Ustilago sativa so as to achieve the site-directed insertion of the Ustilago sativa genome .
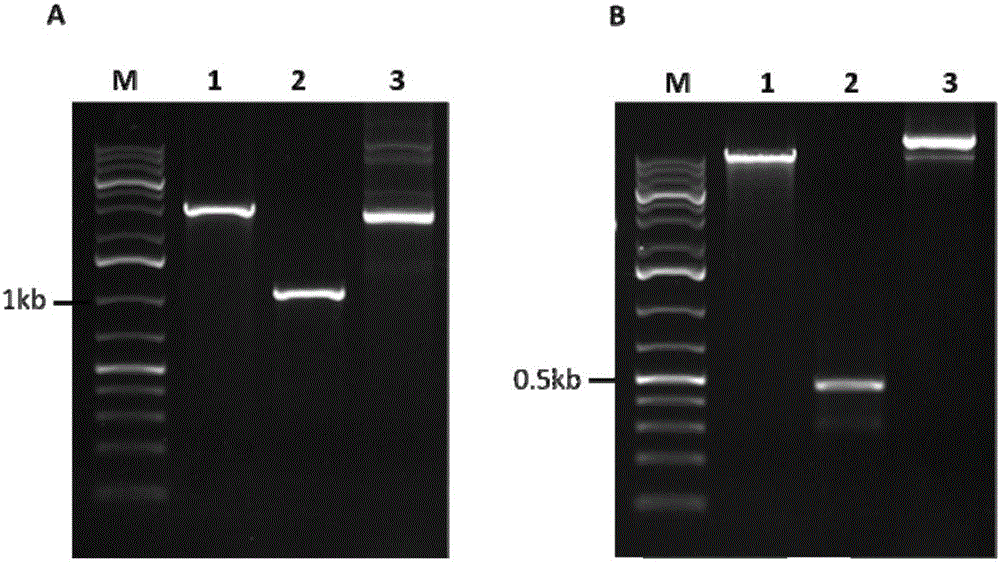
[0026] In order to further clarify the above inventive concept, the sugarcane smut pheromone gene mfa2 is taken as an example for illustration. The experimental methods in the following examples, unless otherwise specified, are conventional methods; the experi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Aperture | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com