Probe, kit and method thereof for detecting MAPT gene mutation
A kit and probe technology, applied in the field of biomedicine, can solve the problems of requiring a large number of manual operations, large human errors, and low degree of automation, and achieve the effects of reducing the probability of cross-contamination, being easy to automate, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
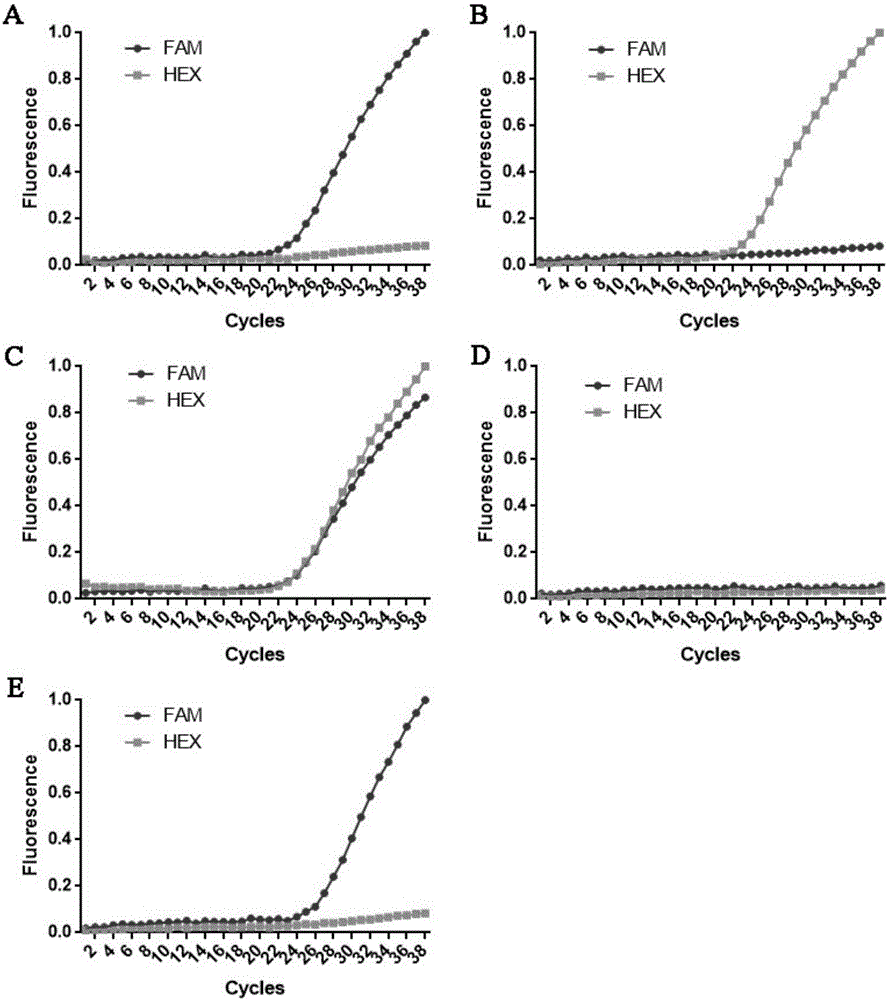
[0043] Example 1 Real-time fluorescent PCR detection of the genotype of the MAPT gene encoding protein R5L mutation site
[0044] 1. Materials
[0045] 1. Instrument:
[0046] Real-time fluorescent PCR instrument, pipette, centrifuge.
[0047] 2. Primer and probe design:
[0048] The present invention designs primers capable of specifically amplifying the R5 site of the protein encoded by the MAPT gene, and probes that specifically recognize the wild type and the mutant type of the R5 site of the protein encoded by the MAPT gene. The 5' end of the wild-type probe is labeled FAM, and the 3' end is labeled BHQ1; the 5' end of the wild-type probe is labeled HEX, and the 3' end is labeled BHQ1.
[0049] The primers and probes used are listed in Table 1.
[0050] Table 1 Primer and probe information list
[0051]
[0052]
[0053] 3. Reagents:
[0054] 1×PCR buffer: 10mmol / L Tris-HCl (pH 8.3), 50mmol / L KCl; MgCl 2 ; Hot start Taq enzyme; dATP, dCTP, dGTP, dTTP.
[005...
Embodiment 2
[0069] Example 2 Real-time fluorescent PCR detection of the genotype of the MAPT gene-encoded protein A152T mutation site
[0070] According to the method and steps of Example 1, real-time fluorescent PCR amplification was performed.
[0071] The primers and probes used are listed in Table 1.
[0072] Result analysis:
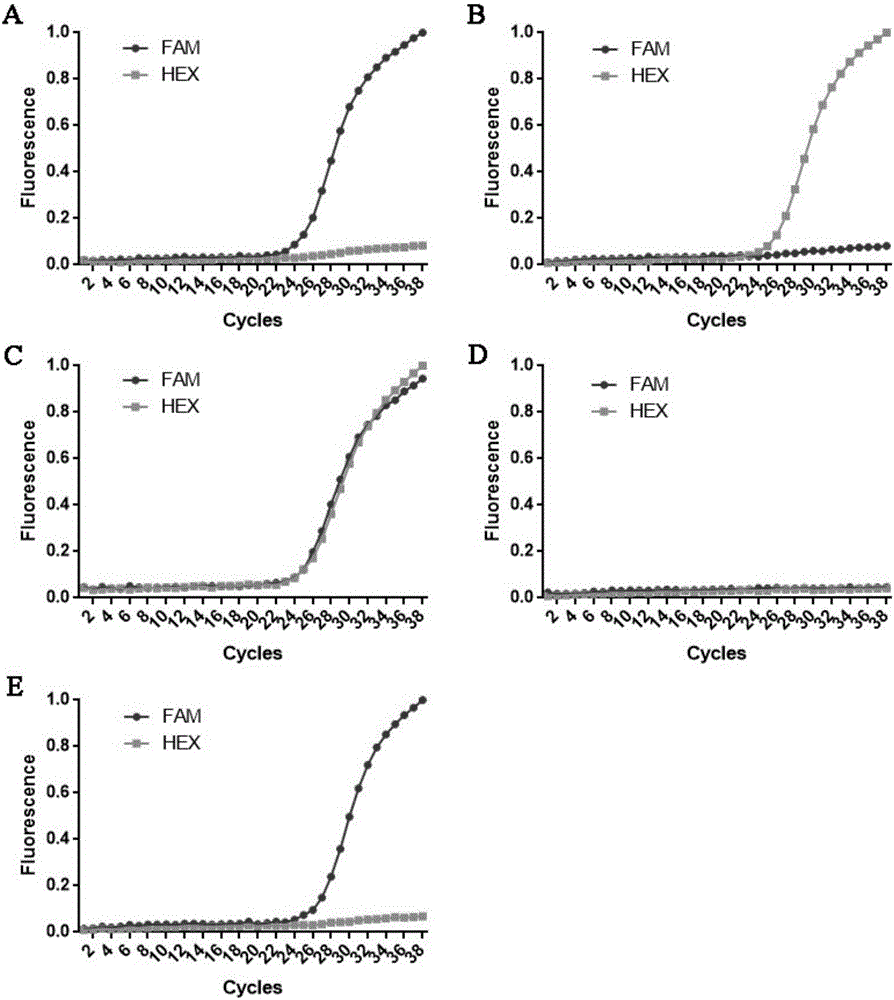
[0073] Genotype determination of the A152 locus of the MAPT gene-encoded protein: samples with amplification signals only in the FAM channel are homozygous for WT / WT; samples with amplification signals only in the HEX channel are homozygous for the A152T / A152T mutation; Samples that produced amplified signals in both the FAM and HEX channels were WT / A152T heterozygotes.
[0074] from figure 2 It can be seen that (A) is the positive standard 1 of the known MAPT genotype, which only has an amplification signal in the FAM channel, and is WT / WT homozygous, which is consistent with the actual situation; (B) is the known MAPT gene Genotype positive standard 2, o...
Embodiment 3
[0075] Example 3 Real-time fluorescent PCR detection of the genotype of the L284R mutation site of the protein encoded by the MAPT gene
[0076] According to the method and steps of Example 1, real-time fluorescent PCR amplification was performed.
[0077] The primers and probes used are listed in Table 1.
[0078] Result analysis:
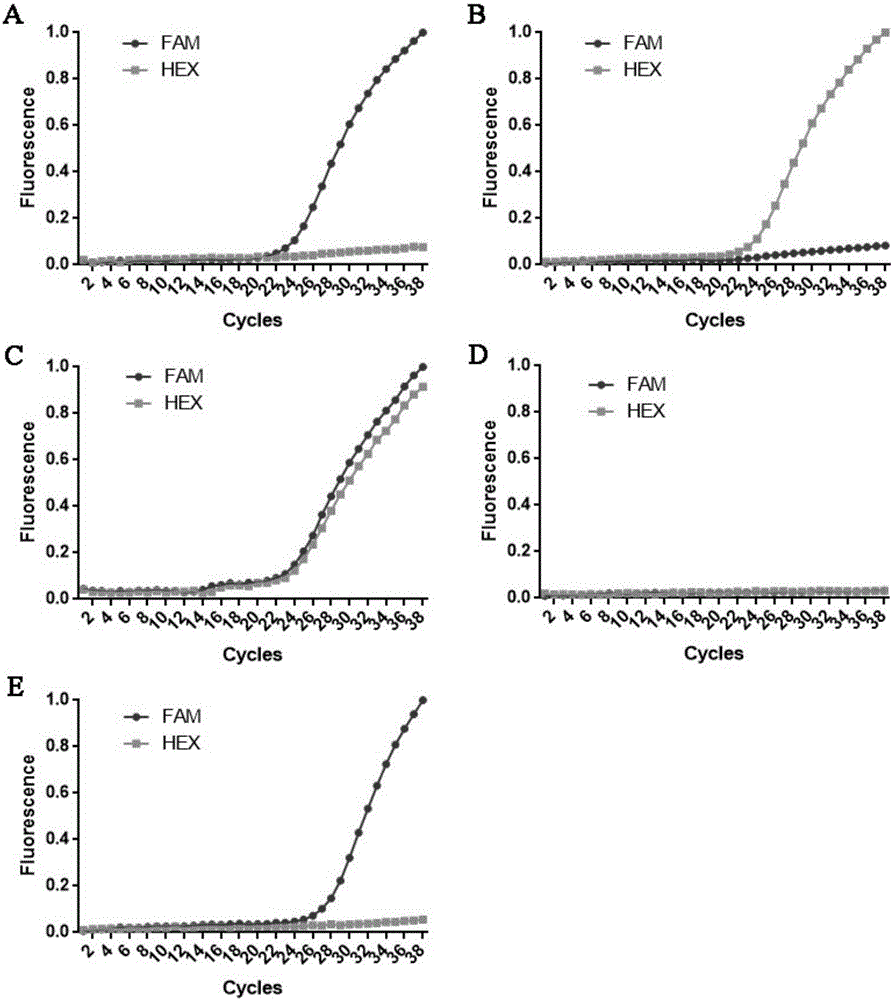
[0079] Genotype determination of the L284 locus of the MAPT gene-encoded protein: samples with amplification signals only in the FAM channel are homozygous for WT / WT; samples with amplification signals only in the HEX channel are homozygous for the L284R / L284R mutation; Samples that produced amplified signals in both the FAM and HEX channels were WT / L284R heterozygotes.
[0080] from image 3 It can be seen that (A) is the positive standard 1 of the known MAPT genotype, which only has an amplification signal in the FAM channel, and is WT / WT homozygous, which is consistent with the actual situation; (B) is the known MAPT gene The positive stand...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com