A method for targeted knockdown of multiple copies of genes in the zebrafish genome
A genome and zebrafish technology, applied in recombinant DNA technology, cells modified by the introduction of foreign genetic material, and introduction of foreign genetic material using vectors, etc., can solve the problem of not finding homologous genes, and achieve a wide range of applications.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] The present invention will be further explained below in conjunction with specific embodiments.
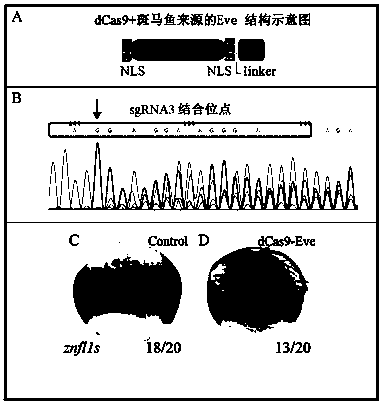
[0022] refer to figure 1 , a method for directional knocking down multi-copy genes in the zebrafish genome proposed by the present invention, comprising the following steps:
[0023] Step 1: Preparation of zebrafish embryos. Zebrafish are kept in a circulating water system at a room temperature of 28.5°C, with 14 hours of light and 10 hours of darkness every day. They are fed once in the morning and in the afternoon. The female fish and the male fish are placed at both ends of the mating box, separated by a partition in the middle. When the light is on the next morning, the partition is pulled out, and the male and female mate. The embryos produced will sink to the bottom of the mating box due to gravity and collected. embryos, and place the embryos in a constant temperature incubator at 28.5°C for future use;
[0024] Step 2: Construction of the expression vector of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com