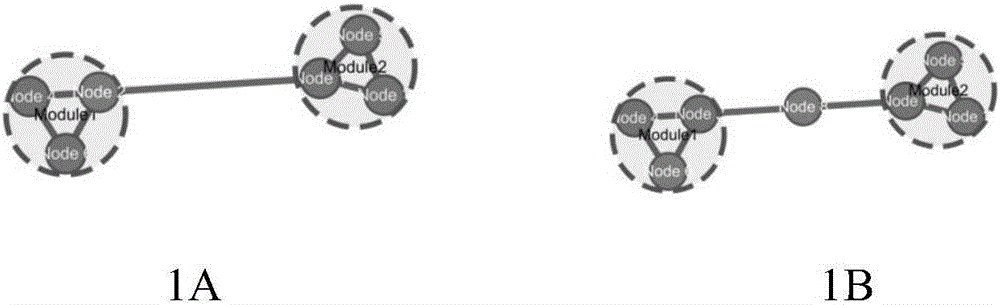
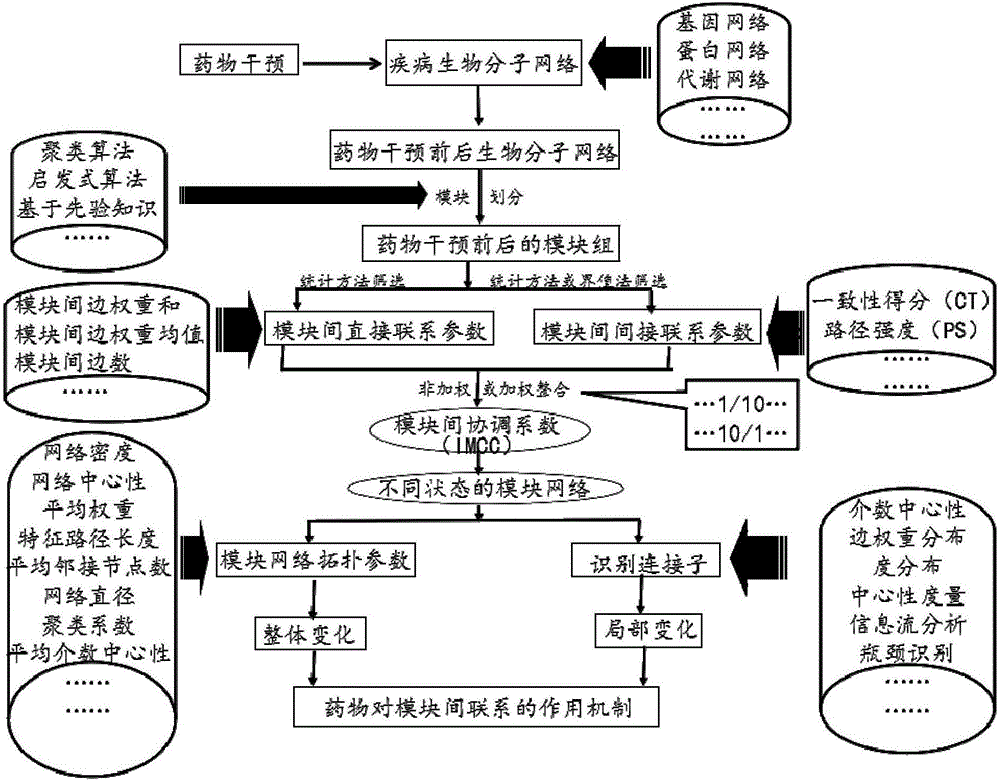
Method for evaluating influences of drugs on inter-module relations in biomolecule network
A biomolecular network and inter-module technology, applied in the field of impact evaluation, can solve the problem of few research methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] This example uses the gene chip experimental data of the effective components of Qingkailing intervening in the mouse cerebral ischemia model as an example, (the data has been uploaded to the ArrayExpress database, Http: / / www.ebi.ac.uk / arrayexpress / ), construct a weighted gene co-expression network, specifically describe the method of the present invention:
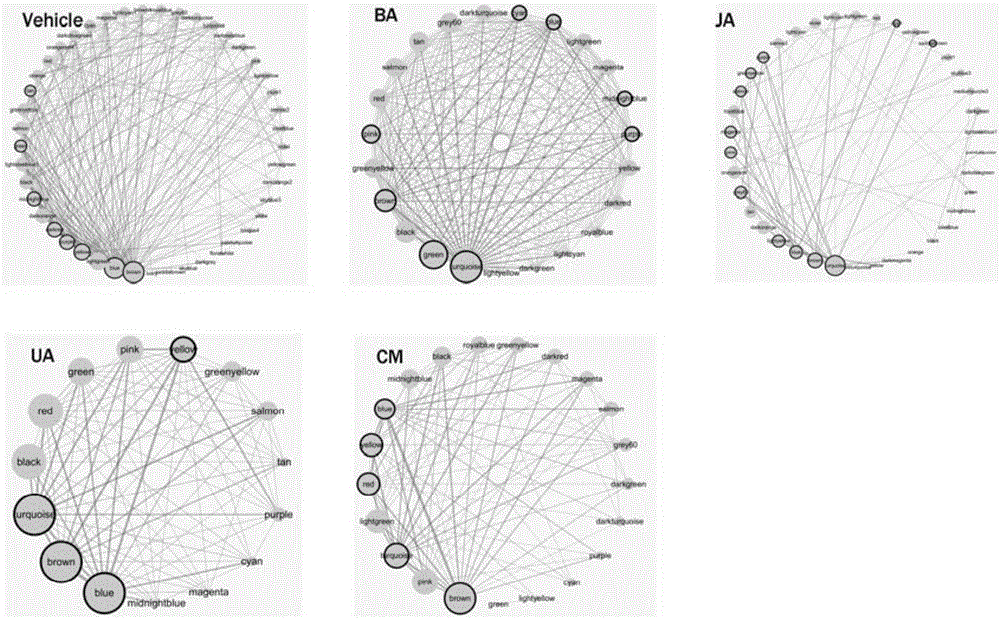
[0081] Based on the weighted gene co-expression network and module division of each component of Qingkailing intervening in the cerebral ischemia mouse model, cerebral ischemia model group (Vehicle), baicalin (BA) group, geniposide (JA) group, cholic acid The module division of (UA) group and mother of pearl (CM) group is shown in Table 3.
[0082] Table 3 Module division of gene co-expression network
[0083]
[0084] Step 1, use the weighted co-expression network analysis (WGCNA) tool (a module division method based on clustering algorithm, source Peter Langfelder, Steve Horvath. WGCNA: an R package for weigh...
Embodiment 2
[0112] Taking the effective components of Qingkailing to intervene in the protein interaction network of the mouse cerebral ischemia model as an example, the method of the present invention is described in detail:
[0113] Based on the protein interaction network and module division of each component of Qingkailing intervening in the cerebral ischemia mouse model, the module division of the cerebral ischemia model group and the baicalin (BA) group (see Zhang Yingying, Qingkailing multi-component intervention Identification and comparison of main modules of protein network in cerebral ischemia model [D], Chinese Academy of Chinese Medical Sciences, 2014), as shown in Table 9.
[0114] Table 9 Module division of protein interaction network
[0115]
[0116] Step 1. By mapping the differential genes before and after the drug intervention to the string protein database, construct the protein interaction network before and after the intervention of each component of Qingkailing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com