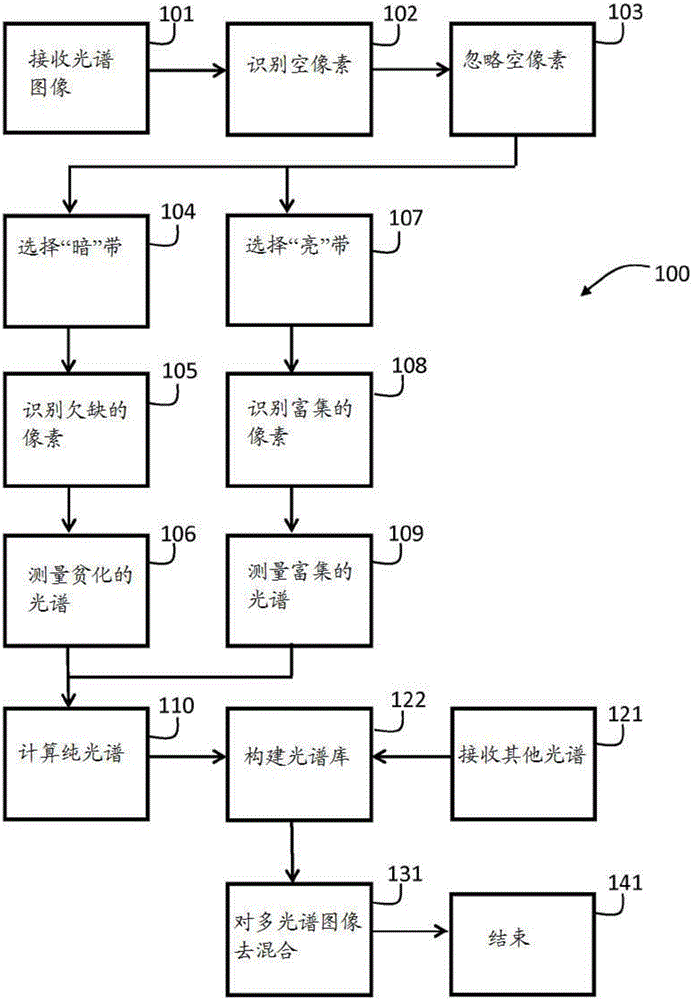
Pure spectrum extraction from biological samples
A purely spectroscopic, sample-based technique used in the imaging of biological samples to address issues such as increased complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0160] In this example, the specimen is a breast cancer specimen obtained as a formalin-fixed, paraffin-enclosed (FFPE) tissue block from which 4-micron sections were dissected using a microtome and subsequently subjected to standard histological procedures. processed for deparaffinization, antigen retrieval, and used in conjunction with The PR probe of 594 dye (available from Life Science Solutions, Caliper, Calif.) was immunofluorescent (IF) labeled. They were counterstained with DAPI and then coverslipped on standard microscope slides. It was imaged using the same tools, ext-filters and wavelengths as previously discussed.
[0161] In this example, the goal is to determine Pure dye spectrum of 594, so dark bands include all image cube planes acquired with DAPI or FITC external filters, and bright bands include all image cube planes acquired with Texas Red filter. A color image is generated from the image cube; the blue, green, and red image planes are shown as Figure...
example 2
[0166] The same procedure as in Example 1 was followed on another sample from the same tissue block on which no DAPI counterstain was applied. Figure 11A The averaged dye-rich, dye-deficient and pure dye spectra obtained are shown as curves 161, 162 and 163, respectively. The dark band was then set to zero and the resulting spectrum was normalized; the result is shown as curve 164 .
[0167] use to measure based on this sample for Spectra of 594 dyes, utilizing the same autofluorescence blank as in the previous example. The same operator performs the same steps as previously described; pure spectra obtained for the dye-loaded, autofluorescent in Figure 11B are shown as curves 165, 166 and 167, respectively.
[0168] Comparing curves 154 and 164, there is no major difference between them, indicating that despite the presence of nuclear-localized (PR antibodies and thus Strong, promiscuous DAPI signal in the same cell compartment in which 594 is mainly located), still ...
example 3
[0171] In a further example, the sample is a breast cancer sample obtained as a formalin-fixed, paraffin-enclosed (FFPE) tissue block from which 4 micron sections were cut using a microtome and subsequently subjected to standard tissue Chemical treatment for dewaxing and antigen retrieval. However, immunofluorescence (IF) calibration was not performed. They were counterstained with DAPI and then coverslipped on standard microscope slides. It was imaged using the same tools, ext-filters and wavelengths as previously discussed.
[0172] Pure DAPI spectra were determined according to the method disclosed herein, using all image cube layers for which FITC external filters were utilized as dark bands. Bright bands include all image cube layers for which the DAPI extrinsic filter was utilized. Image cube layers acquired with the Texas Red extrinsic filter are not assigned to dark or light bands. Dye-rich and dye-deficient pixels were selected using the same procedure and thresho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com