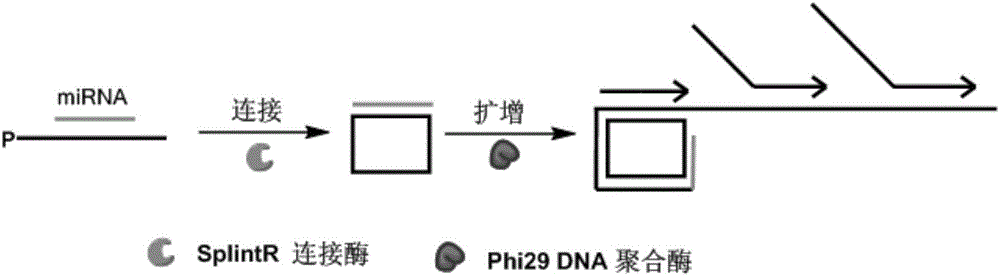
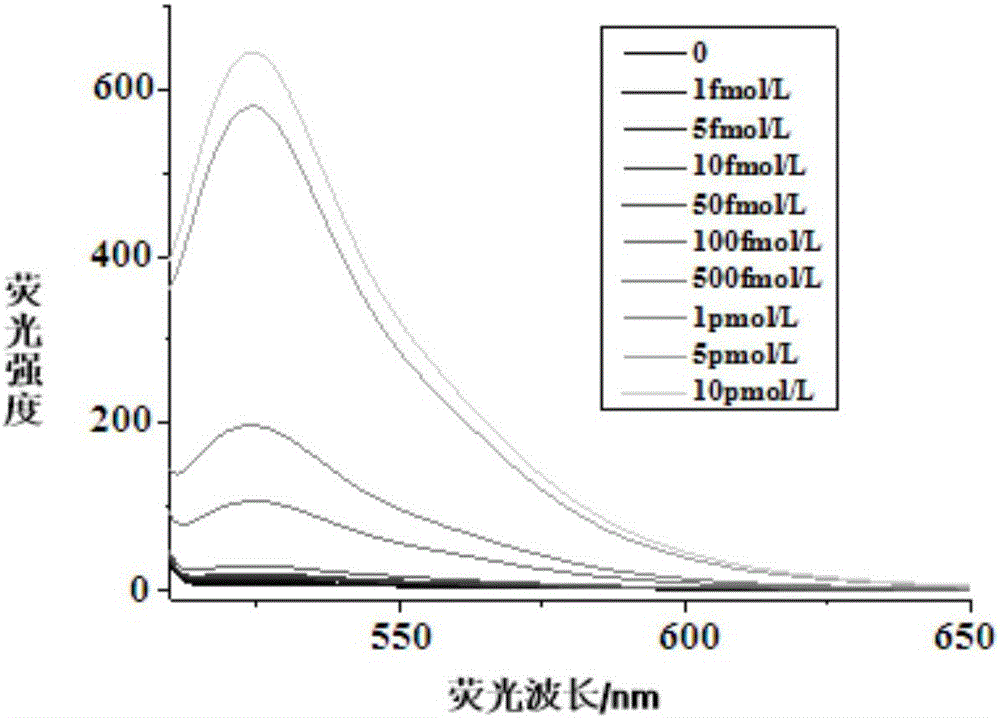
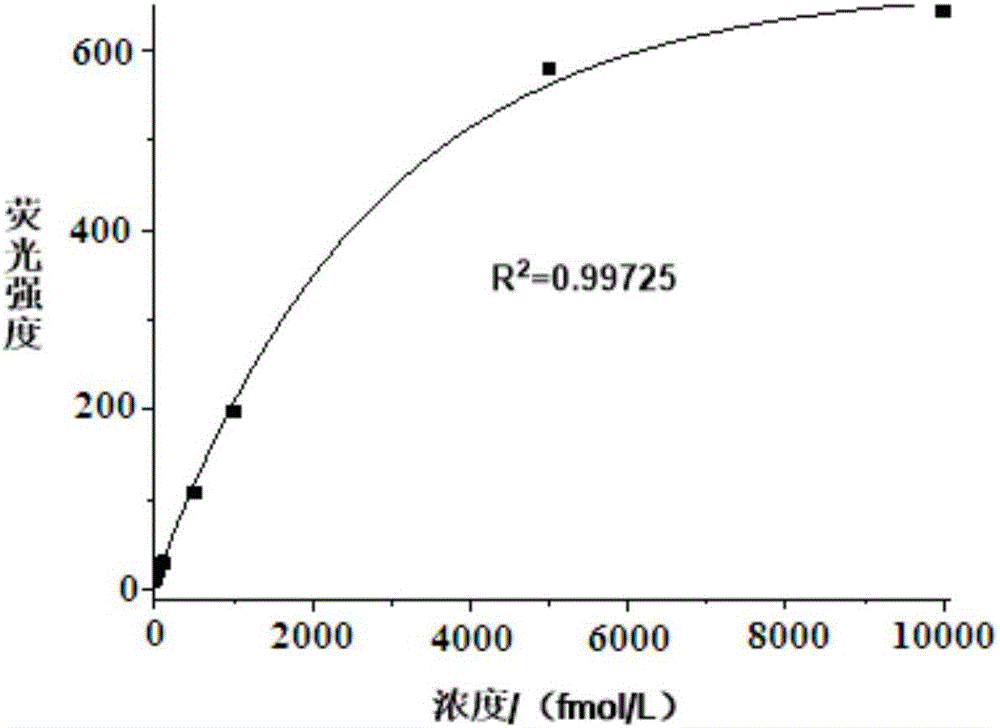
Method for detecting miRNA by ultra-branch rolling circle amplification triggered by connection reaction
A technology of rolling circle amplification and ligation reaction, applied in the fields of molecular biology and nucleic acid chemistry, which can solve problems such as unsatisfactory ligation efficiency and impact on detection sensitivity, so as to avoid the interference of background fluorescent signals, improve ligation efficiency, and improve sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] In this example, miRNA21 (sequence 5'-uagcuuaucagacugauguuga-3') was used as the detection target.
[0027] 1. Design and synthesis of 5' phosphorylated padlock probe P-21 and secondary primer S-1
[0028] (1) The sequence of the phosphorylated padlock probe P-21 at the 5' end is:
[0029] 5'- CTGATAAGCTA TTTGCATTTCAGTTTACGGTTTAGCATTTCGCAATTTT TCAACATCAGT -3'.
[0030] Among them, the underlined sequences at both ends can be complementary paired with miRNA21 (miR-21), respectively, and there is a sequence in the middle with a length of 38 bases.
[0031] (2) The sequence of the secondary primer S-1 is: 5'-AGTCTGATAAGCTA TTTGCA-3', which is identical to the partial sequence of the phosphorylated lock probe, and can be complementary to the linear rolling circle amplification product, thereby triggering hyperbranched rolling circle amplification. increase.
[0032] 2. Detection of miRNA21
[0033] (1) Ligation reaction
[0034] In 10μL ligation reaction system, ad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com