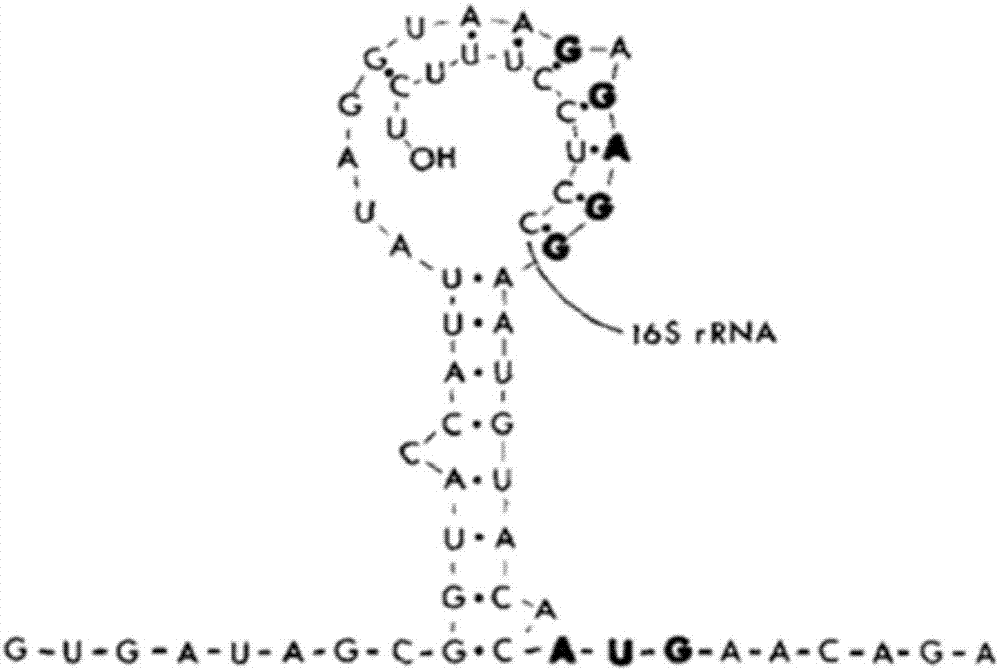
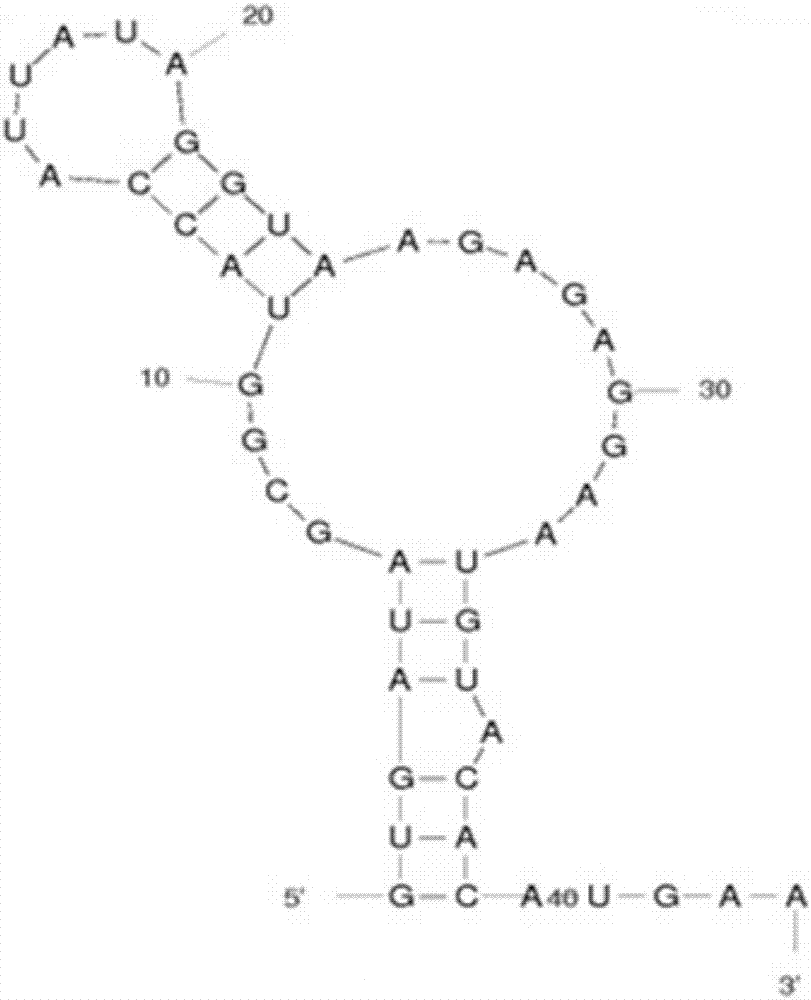
Ribosome binding site reconstruction-based promoter optimization method
A technology of binding sites and optimization methods, applied in the fields of genetic engineering and microbial engineering, can solve problems such as increased difficulty in ribosome recognition, decreased protein expression level, and decreased translation initiation efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] S1. According to the B. subtilis 168 genome sequence NZ_CP010052.1 released by the NCBI database, the original sequence of P43 was found. For the specific sequence, see the sequence listing SEQ ID NO: 1; Primer Primer 5 software was used to design primers P43-F and P43-SOE-R, The original promoter P43 was amplified using the total DNA of B. subtilis 168 as a template;
[0049] Primers were designed as follows:
[0050] P43-F: CGGAATTCTGATAGGTGGTATGTTTTCG
[0051] P43-SOE-R: CTTACCTATAATGGTACCAGATCTGCTATCACTTTAT
[0052] S2. Using the promoter P43 amplified in step S1 as a template, use the Primer Primer5 software to design primers P43-F and P43RBS1-SOE-R, and when designing the direction primer, subtract the 5 bases behind the primer P43RBS1-SOE-R and add the 5 bases of TTCAT in front of the primer P43RBS1-SOE-R, the promoter RBS1 is amplified by PCR, so that the sequence ATGAA is more behind the amplified sequence, and its specific sequence is shown in the sequence t...
Embodiment 2
[0058] S1. According to the B. subtilis 168 genome sequence NZ_CP010052.1 released by the NCBI database, the original sequence of P43 was found. For the specific sequence, see the sequence listing SEQ ID NO: 1; Primer Primer 5 software was used to design primers P43-F and P43-SOE-R, The original promoter P43 was amplified using the total DNA of B. subtilis 168 as a template;
[0059] Primers were designed as follows:
[0060] P43-F: CGGAATTCTGATAGGTGGTATGTTTTCG
[0061] P43-SOE-R:
[0062] CTTACCTATAATGGTACCAGATCTGCTATCACTTTAT;
[0063] S2. Using the promoter P43 amplified in step S1 as a template, use the Primer Primer5 software to design primers P43-F and P43RBS2-SOE-R, remove the two bases C and A from the P43 neck loop structure, and remove the RBS The binding region is changed from GTAAGAGAGG to GAAAGGAGG, and the promoter RBS2 is amplified by PCR, and its specific sequence is shown in SEQ ID NO:3 in the sequence table.
[0064] Primers were designed as follows:
[0...
Embodiment 3
[0069] S1. According to the B. subtilis 168 genome sequence NZ_CP010052.1 released by the NCBI database, the original sequence of P43 was found. For the specific sequence, see the sequence listing SEQ ID NO: 1; Primer Primer 5 software was used to design primers P43-F and P43-SOE-R, The original promoter P43 was amplified using the total DNA of B. subtilis 168 as a template;
[0070] Primers were designed as follows:
[0071] P43-F: CGGAATTCTGATAGGTGGTATGTTTTCG
[0072] P43-SOE-R:
[0073] CTTACCTATAATGGTACCAGATCTGCTATCACTTTAT;
[0074] S2. Using the promoter P43 amplified in step S1 as a template, use the Primer Primer5 software to design primers P43-F and P43RBS2-SOE-R, remove the two bases C and A from the P43 neck loop structure, and remove the RBS The binding region was changed from GTAAGAGAGG to GAAAGGAGG, and the promoter RBS2 was amplified by PCR;
[0075] Primers were designed as follows:
[0076] P43-F: CGGAATTCTGATAGGTGGTATGTTTTCG
[0077] P43RBS2-SOE-R:
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com